CABBI researchers in Feedstock Production, Conversion, and Sustainability develop and modify models to determine efficient, cost-effective methods to grow bioenergy grasses, convert them to useful chemicals, and positively impact both the bioeconomy and the environment.
The details for the models listed on this page — divided into categories such as Ecosystem, Design and Economic, Plant Physiology, and Metabolic — include contact information for the scientists who use them.
Some of the models are publicly available; please follow the links in the descriptions to access these.
Ecosystem Models
Researcher: Andy VanLoocke
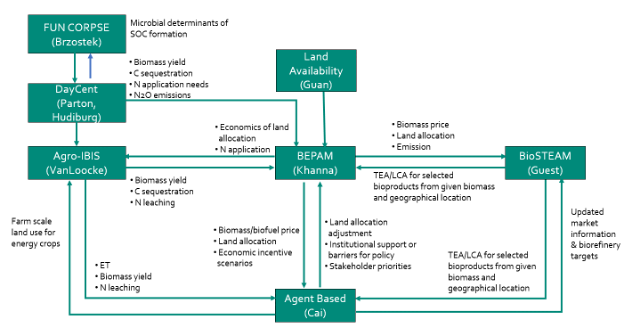
Agro-IBIS (Integrated Biosphere Simulator – Agricultural Version) is a process-based agroecosystem model. It simulates energy, water, carbon, and nitrogen exchange between the soil-plant-atmosphere continuum by simulating biophysical cycles and biochemical processes on an hourly time step.
Within CABBI, Agro-IBIS is being used in conjunction with other models including BEPAM to understand water quality impacts and nitrogen delivery in bioenergy cropping systems.
Related data:
Researchers: William Parton, Melannie Hartmann, Elena Blanc-Betes

DayCent (Daily Century model) is a biogeochemical model that simulates fluxes of carbon and nitrogen among the atmosphere, vegetation, and soil to produce daily fluxes of nitrogen gases, CO2 flux from heterotrophic soil respiration, soil organic carbon and nitrogen, net primary production, and H2O and NO3– leaching.
CABBI researchers have modified DayCent to add a “GrassTree” crop type to better represent large bioenergy crops and grasses in the model. These can be either annual or perennial and leaves and stems can be managed separately, so representing crop harvest is more flexible.
To access the source code for the bioenergy crop related updates, please see the DayCent-CABBI data webpage. For more information about the general DayCent model, see the DayCent website.
Related data:
Researchers: Edward Brzostek, Stephanie Juice
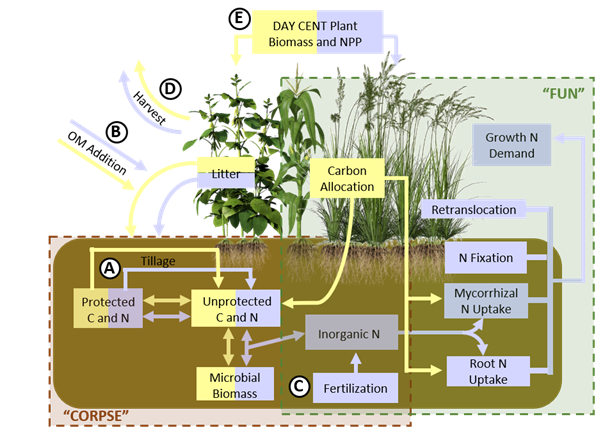
FUN-BioCROP (Fixation and Uptake of Nitrogen — Bioenergy, Carbon, Rhizosphere Organisms and Protection), is modified from a coupling of the FUN model that predicts the allocation of carbon to the rhizosphere and the CORPSE model that predicts the impacts of the plant carbon on soil carbon and nitrogen cycling to model bioenergy, plant-microbial interactions.
Changes implemented by CABBI researchers include developing crop-specific parameters to characterize soil carbon dynamics differences, adding tillage parameters to make carbon accessible to microbial decomposition, and having harvests reduce the plant litter added to soil pools. The model is still under development and not yet publicly available.
Related Data:
Design and Economic Models
Researchers: Jeremy Guest, Yoel Cortés-Peña, Yalin Li
BioSTEAM is an open-source Python package for the design, simulation, and techno-economic analysis of biorefineries under uncertainty. By incorporating uncertainty and sensitivity analyses as a key feature, BioSTEAM enables the evaluation of complete landscapes of design decisions for established, emerging, and conceptualized technologies under a broad range of scenarios (e.g., policy frameworks, locations, market prices). BioSTEAM allows users to build new unit operation models and share designs transparently and without economic barriers.
BioSTEAM-LCA interfaces with the BioSTEAM to provide agile life cycle analysis to streamline and automate early-stage environmental impact analyses of processes and technologies, and to enable rigorous sensitivity and uncertainty analyses linking process design, performance, economics, and environmental impacts. It allows for the assessment of environmental and economic sustainability of biorefinery production processes for a range of different design choices.
Related data:
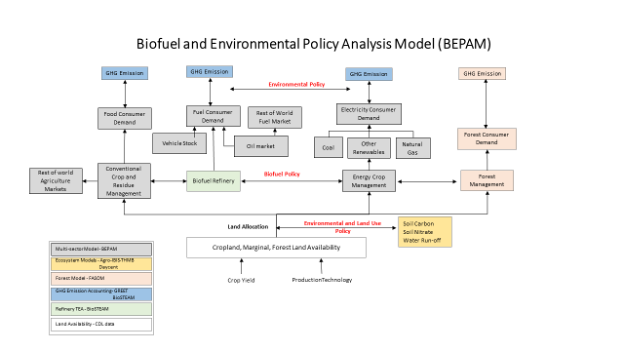
Researcher: Madhu Khanna
BEPAM (Biofuel and Environmental Policy Analysis Model) is a multi-period, open-economy, spatial, multi-market optimization model that analyzes the market decisions on land allocation, crop mix, biofuel feedstock mix, spatial locations of production for various row crops and energy crops in response to various market and policy scenarios.
CABBI researchers are linking BEPAM to ecosystem models, refinery scale techno-economic analysis and life-cycle emissions accounting to determine the interactions among policy-induced land use changes, economic costs and benefits, greenhouse gas mitigation, soil carbon sequestration and water quality.
Related data:
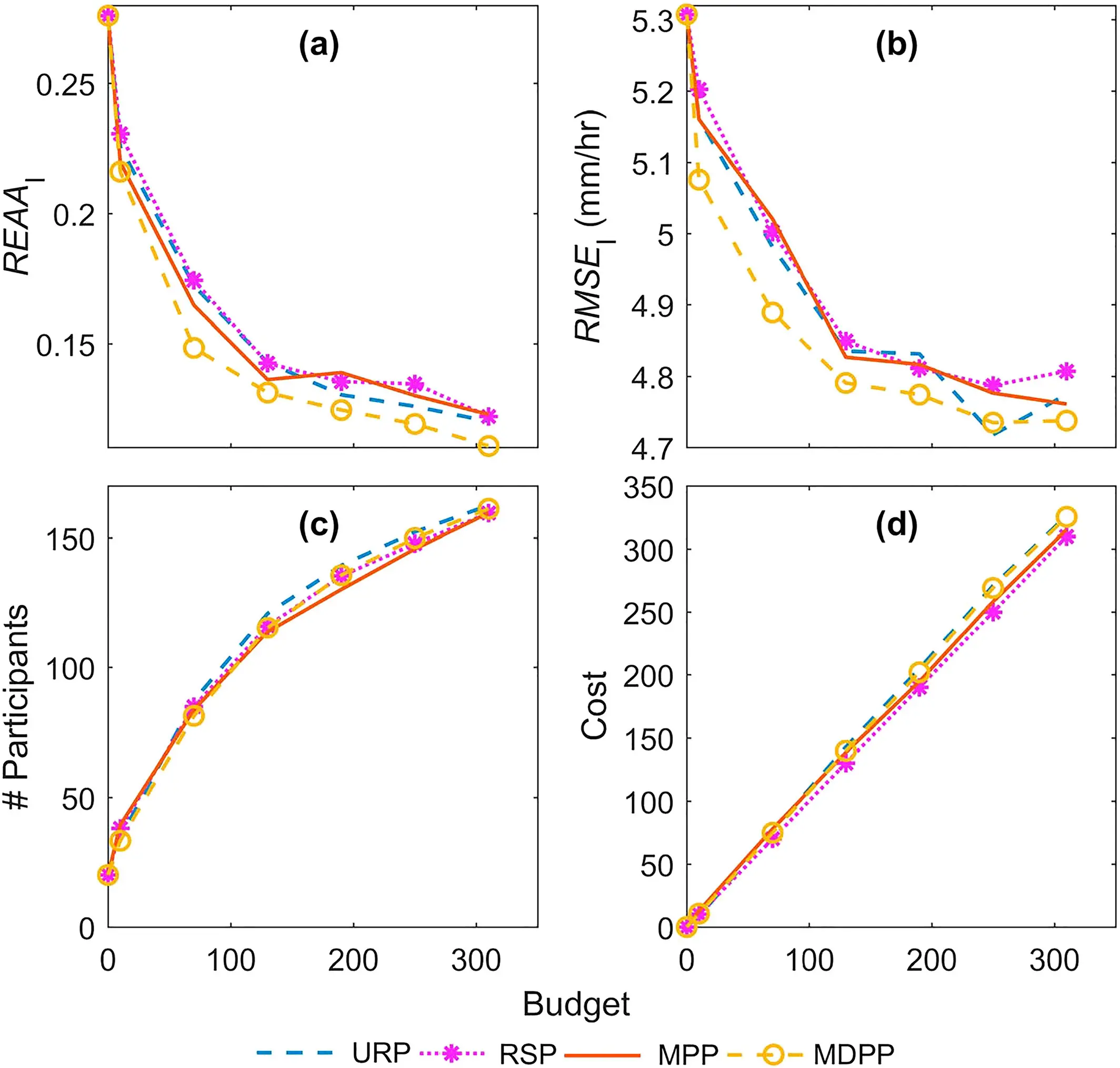
Researcher: Ximing Cai
The CABBI Reward Allocation — Agent-Based Model (RA-ABM) was developed to represent the decision-making processes for a central manager in determining reward allocation to maximize crowdsourcing participation for environmental monitoring. This agent-based model simulates the interactions between the manager and participants to evaluate the effectiveness of reward allocation policies.
Related data:
Plant Physiology Models
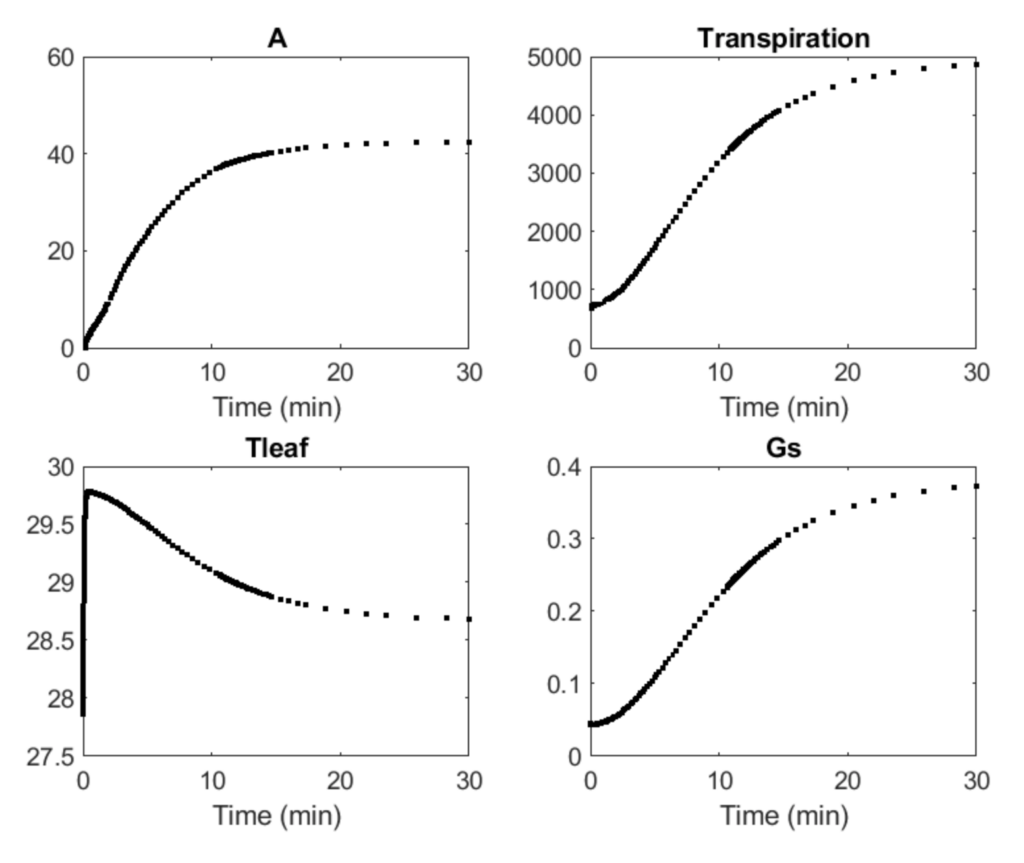
Researchers: Stephen Long, Yu Wang
The C4 Dynamic Model, a dynamic systems model of C4 crop photosynthesis, is based on the NADP-ME metabolic model for maize, an ordinary differential equation model that includes individual steps in C4 photosynthetic carbon metabolism. The NADP-ME is extended to include posttranslational regulation and temperature response of enzyme activities, dynamic stomata conductance, and leaf energy balance.
Related data:
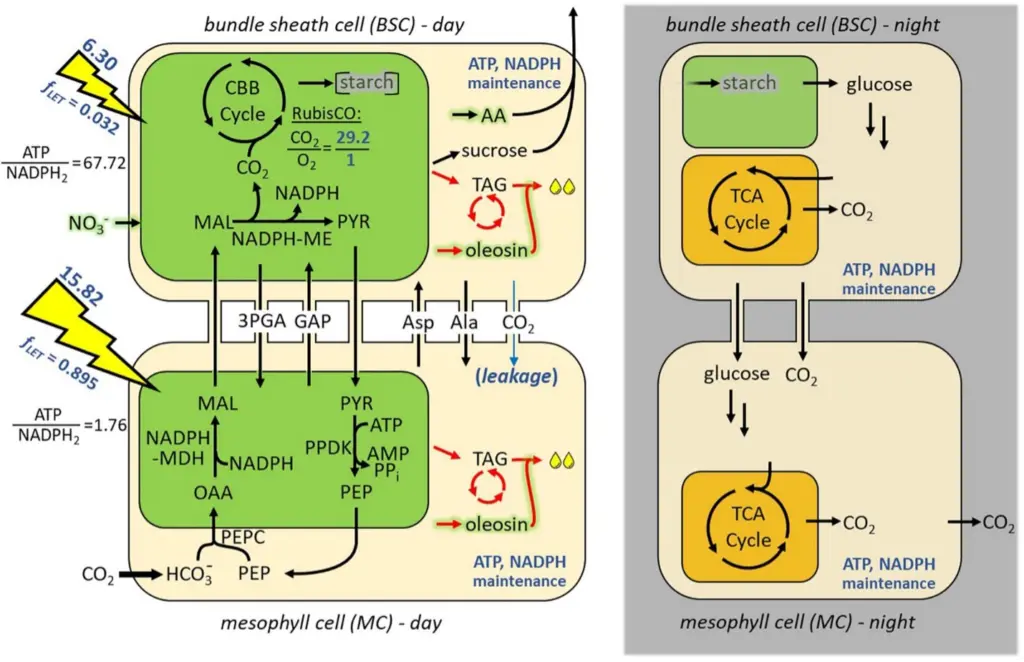
Researchers: Jörg Schwender, Teresa Clark
The iTJC1414 model is a genome-referenced core metabolic model of Sorghum bicolor with detailed manual curation of NADP-malic enzyme (NADP-ME) type C4 photosynthesis, lipid metabolism and other parts of central metabolism. A two-cell leaf model representing C4 metabolism was constructed to investigate the metabolic potential to photo-assimilate CO2 into carbohydrate or TAG. This was then expanded into a diel model that simulates cycles of day and night leaf metabolism (iTJC1414x4). The metabolic reaction network is based on iEB2140x2, a two-cell high confidence representation of Zea mays core metabolism.
Related data:
Metabolic Models
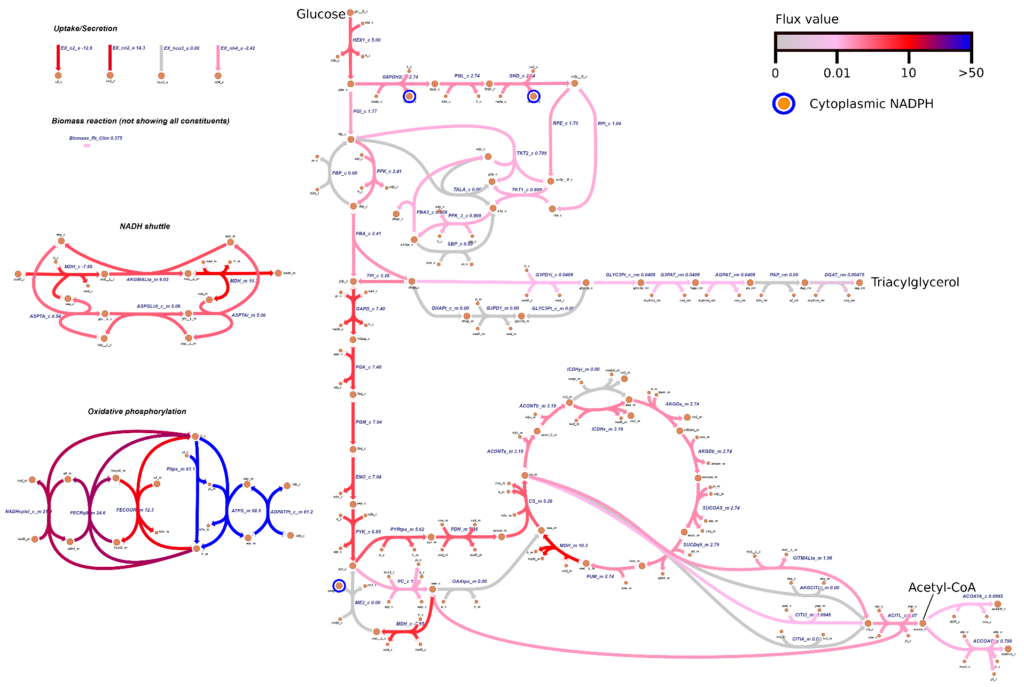
Researcher: Costas Maranas
iRhto1108 is a genome-scale model of Rhodosporidium toruloides IFO0880’s metabolic network. It integrates yeast biochemistry information from previous research and new data to account for 2204 reactions, 1985 metabolites, and 1108 genes. The included genes cover 13% of the organism’s chromosomal and 6% of mitochondrial genome.
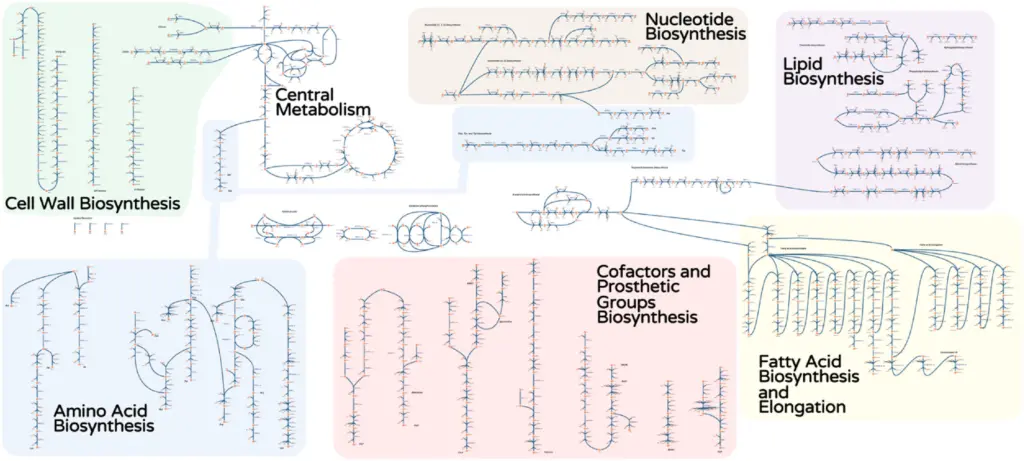
Researchers: Costas Maranas, Patrick Suthers
iIsor850 is a genome-scale metabolic model for Issatchenkia orientalis SD108 that assesses the metabolic capabilities of this non-model yeast. It spans 850 genes, 1826 reactions and 1792 metabolites. Initially, based on mapping genes and reactions from the Saccharomyces cerevisiae genome-scale model Yeast 7.6, the model is refined with other studies and laboratory work.
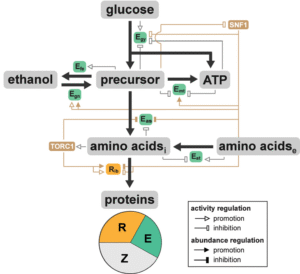
Researcher: Ting Lu
This coarse-grained, systems, and dynamic mathematical framework model was developed to determine the emergent metabolic traits of Saccharomyces cerevisiae by integrating metabolic reactions with signal transduction and gene regulation. The framework captures a set of characteristic cellular behaviors, including the Crabtree effect, diauxic shift, diauxic lag time, and differential growth under nutrient-altered environments and allows modular expansion for zooming in on specific pathways for detailed metabolic profiles.
