C4 Dynamic Model and Gas Exchange Data
Themes: Feedstock Production
Keywords: Modeling, Software
Citation
Model Citation
Wang, Y., Chan, K.X., Long, S.P. March 2021. C4 Dynamic Model, GitHub Repository, https://github.com/yuwangcn/C4_dynamic_model.
Data Citation
Wang, Y., Chan, K.X., Long, S.P. May, 5 2021. “Data for ‘Towards a Dynamic Photosynthesis Model to Guide the Yield Improvement in C4 Crops.’ ” University of Illinois Urbana-Champaign. DOI: 10.13012/B2IDB-2694900_V1.
Overview
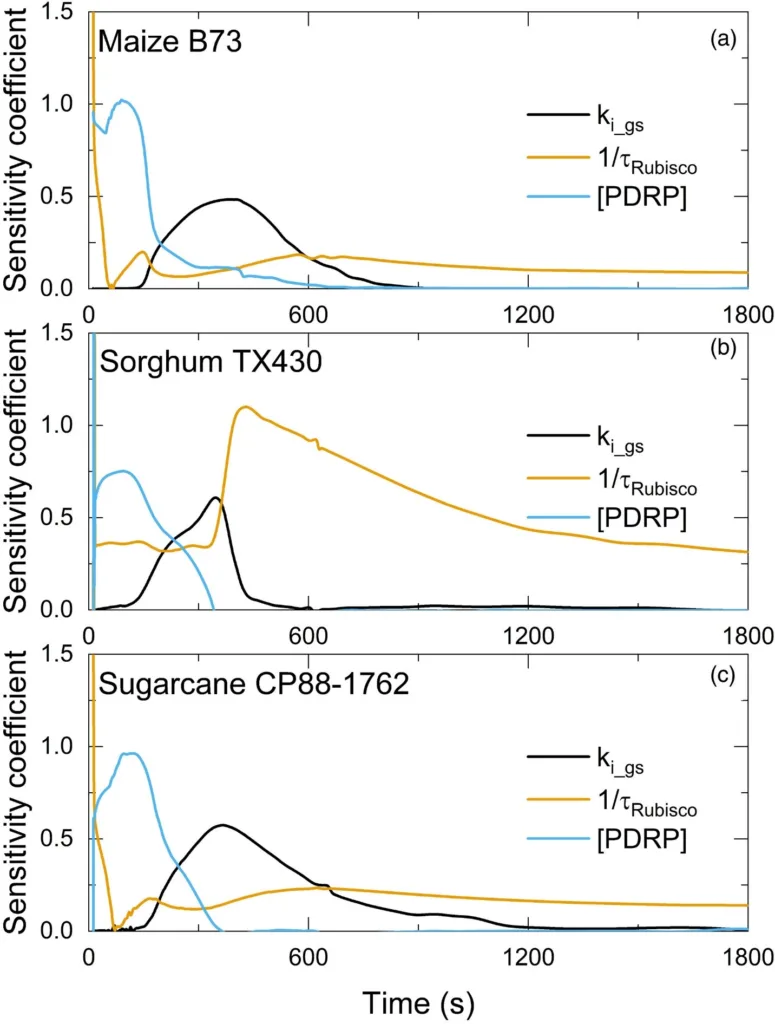
A dynamic systems model of C4 photosynthesis was developed based on the previous NADP-ME metabolic model for maize (Wang et al., 2014 ab). The NADP-ME metabolic model is an ordinary differential equation model including all individual steps in C4 photosynthetic carbon metabolism. Here, the model is extended to include posttranslational regulation and temperature response of enzyme activities, dynamic stomata conductance, and leaf energy balance. This model is written in Matlab (R2019a)
Steady-state and dynamic gas exchange data for maize (B73), sugarcane (CP88-1762) and sorghum (Tx430) were measured in a greenhouse in Urbana, IL from July 25, 2019 through August 8, 2019. Data include:
- CO2 response curves
- Light response curves
- Photosynthetic induction curves measured in the transition from darkness to high light (1800 μmol m-2 s-1), data logged every 1 min.
- Photosynthetic induction in the transition from darkness to high light (1800 μmol m-2 s-1) to determine the kinetics of rubisco activation in these C4 crops (τ_Rubisco), data logged every 10 s.
- Gas exchange under fluctuating light. After dark adaptation, the leaves undergo three light change steps, light intensity was set as 1800 µmol m-2 s-1, 200 µmol m-2 s-1 and 1800 µmol m-2 s-1 for each 1800 s step.
Data
Data Download (1.1MB)