System Analysis of Lipomyces starkeyi During Growth on Various Plant‑Based Sugars
Themes: Conversion
Keywords: Metabolomics, Transcriptomics
Citation
Deewan, A., Liu, J.J., Jagtap, S.S., Yun, E.J., Walukiewicz, H., Jin, Y.S., Rao, C.V. July 30, 2022. “System Analysis of Lipomyces starkeyi During Growth on Various Plant-Based Sugars.” Applied Microbiology and Biotechnology. DOI: 10.1007/s00253-022-12084-w.
Overview
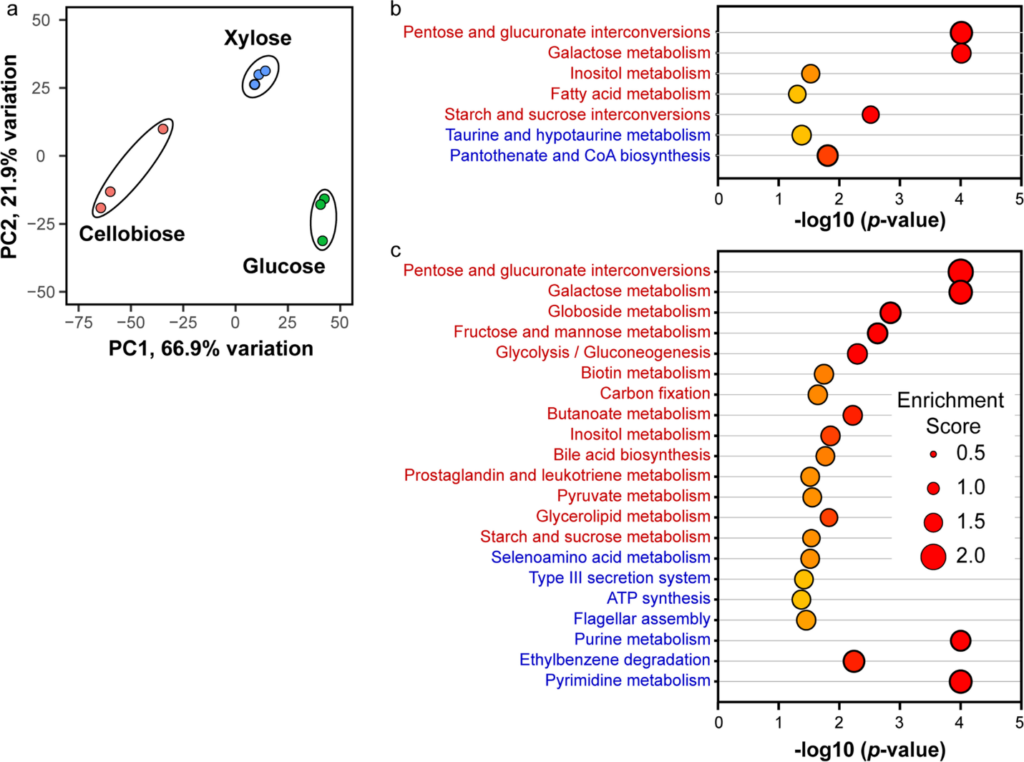
Oleaginous yeasts have received significant attention due to their substantial lipid storage capability. The accumulated lipids can be utilized directly or processed into various bioproducts and biofuels. Lipomyces starkeyi is an oleaginous yeast capable of using multiple plant-based sugars, such as glucose, xylose, and cellobiose. It is, however, a relatively unexplored yeast due to limited knowledge about its physiology. In this study, we have evaluated the growth of L. starkeyi on different sugars and performed transcriptomic and metabolomic analyses to understand the underlying mechanisms of sugar metabolism. Principal component analysis showed clear differences resulting from growth on different sugars. We have further reported various metabolic pathways activated during growth on these sugars. We also observed non-specific regulation in L. starkeyi and have updated the gene annotations for the NRRL Y-11557 strain. This analysis provides a foundation for understanding the metabolism of these plant-based sugars and potentially valuable information to guide the metabolic engineering of L. starkeyi to produce bioproducts and biofuels.
Data
Lipomyces starkeyi RNA Sequencing
Analysis Scripts and Input Data
Download (565KB) includes:
- Differential Gene Expression
- Metabolite List
- List of Annotated Genes