Quantifying the Propagation of Parametric Uncertainty on Flux Balance Analysis
Themes: Conversion
Keywords: Metabolomics, Modeling
Citation
Dinh, H.V., Sarkar, D., Maranas, C.D. Oct. 27, 2021. Data from: “Quantifying the Propagation of Parametric Uncertainty on Flux Balance Analysis.” GitHub Repository.
Overview
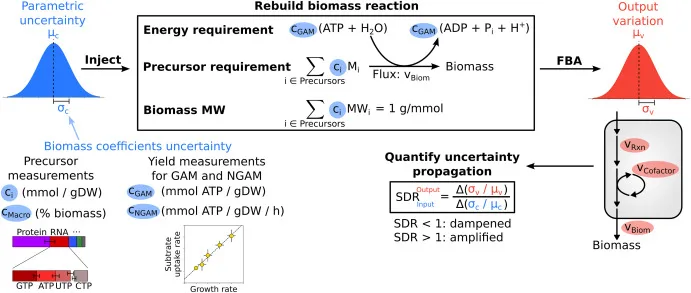
In the repository are example scripts that perform uncertainty injection and propagation to flux balance analysis with outputs for a small sample size (for demonstration purpose only). For proper analysis, user should download the scripts and run for a large sample size (e.g., 10,000 samples).
If you use the scripts, please cite the following Metabolic Engineering article: “Quantifying the propagation of parametric uncertainty on flux balance analysis” (https://doi.org/10.1016/j.ymben.2021.10.012)
There are two subdirectories:
- /uncFBA/uncBiom: injection of normally distributed noise to biomass precursor coeffcients and ATP maintenance (growth-associated ATP maintenance (GAM) and non-growth associated ATP maintenance (NGAM))
- /uncFBA/uncRHS: departure from steady-state by adding noise drawn from normal distribution to the RHS terms of mass balance constraints
Data
GitHub Repository – Includes model scripts.
- Co-factor fluxes
- SDR values
- Alternative metabolic pathways