Quantifying the Propagation of Parametric Uncertainty on Flux Balance Analysis
Themes: Conversion
Keywords: Metabolomics, Modeling
Citation
Dinh, H.V., Sarkar, D., Maranas, C.D. Oct. 27, 2021. Data from: “Quantifying the Propagation of Parametric Uncertainty on Flux Balance Analysis.” GitHub Repository. DOI: 10.11578/dc.20241105.5
Overview
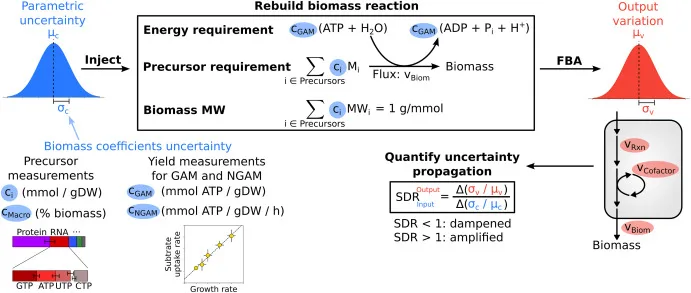
In the repository are example scripts that perform uncertainty injection and propagation to flux balance analysis with outputs for a small sample size (for demonstration purpose only). For proper analysis, user should download the scripts and run for a large sample size (e.g., 10,000 samples).
If you use the scripts, please cite the following Metabolic Engineering article: “Quantifying the propagation of parametric uncertainty on flux balance analysis” (https://doi.org/10.1016/j.ymben.2021.10.012)
There are two subdirectories:
- /uncFBA/uncBiom: injection of normally distributed noise to biomass precursor coeffcients and ATP maintenance (growth-associated ATP maintenance (GAM) and non-growth associated ATP maintenance (NGAM))
- /uncFBA/uncRHS: departure from steady-state by adding noise drawn from normal distribution to the RHS terms of mass balance constraints
Data
GitHub Repository – Includes model scripts.
Illinois Data Bank includes:
- Co-factor fluxes
- SDR values
- Alternative metabolic pathways