Proteotoxicity from Aberrant Ribosome Biogenesis Compromises Cell Fitness
Themes: Conversion
Keywords: Proteomics, RNA Sequencing
Citation
Tye, B.W., Commins, N., Ryazanova, L.V., Wühr, M., Springer, M., Pincus, D., Churchman, L.S. March 7, 2019. Data from: “Proteotoxicity from Aberrant Ribosome Biogenesis Compromises Cell Fitness.” NCBI – Gene Expression Omnibus.
Overview
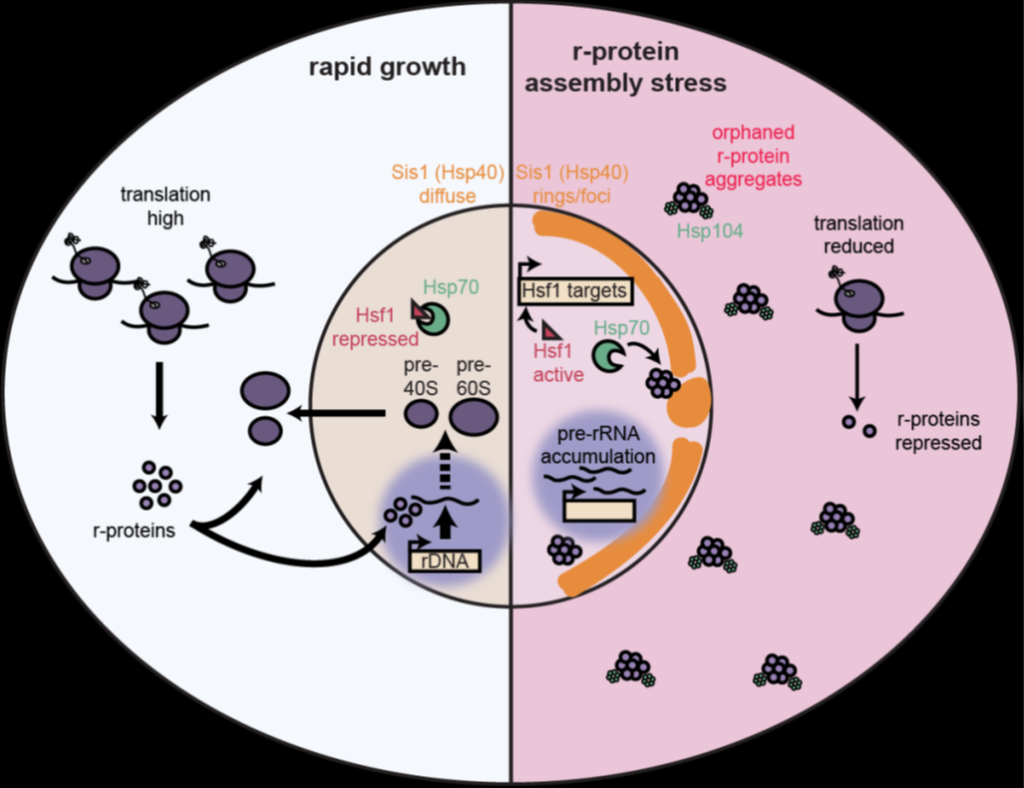
To achieve maximal growth, cells must manage a massive economy of ribosomal proteins (r-proteins) and RNAs (rRNAs) to produce thousands of ribosomes every minute. Although ribosomes are essential in all cells, natural disruptions to ribosome biogenesis lead to heterogeneous phenotypes. Here, we model these perturbations in Saccharomyces cerevisiae and show that challenges to ribosome biogenesis result in acute loss of proteostasis. Imbalances in the synthesis of r-proteins and rRNAs lead to the rapid aggregation of newly synthesized orphan r-proteins and compromise essential cellular processes, which cells alleviate by activating proteostasis genes. Exogenously bolstering the proteostasis network increases cellular fitness in the face of challenges to ribosome assembly, demonstrating the direct contribution of orphan r-proteins to cellular phenotypes. We propose that ribosome assembly is a key vulnerability of proteostasis maintenance in proliferating cells that may be compromised by diverse genetic, environmental, and xenobiotic perturbations that generate orphan r-proteins.
Data
NCBI – Gene Expression Omnibus
- Gene Accession Number GSE114077