Phylogenetic Diversity of Light-dependent Phosphorylation of Thr78 in Rubisco Activase
Themes: Feedstock Production
Keywords: Photosynthesis, Sorghum
Citation
Bhatnagar, N., Chung, S.S., Hodge, J., Kim, S.Y., Sands, M., Leakey, A.D.B., Ort, D.R., Burgess, S.J. July 9, 2025. Data from: “Phylogenetic Diversity of Light Dependent Phosphorylation of Thr78 in Rubisco Activase.” University of Illinois. DOI: 10.13012/B2IDB-4860986_V1.
Overview
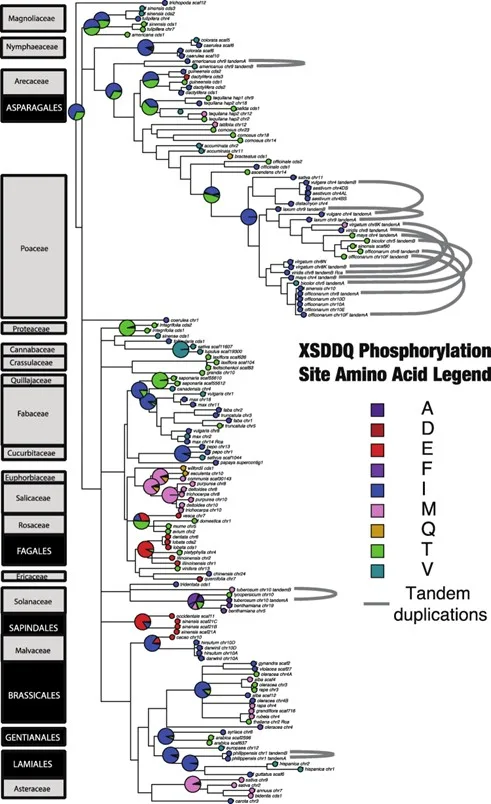
Rubisco activase is an ATP-dependent chaperone that facilitates dissociation of inhibitory sugar phosphates from the catalytic sites of Rubisco during photosynthesis. In Arabidopsis, Rubisco activase is negatively regulated by dark-dependent phosphorylation of Thr78. The prevalence of Thr78 in Rubisco activase was investigated across sequences from 91 plant species, finding that 29 (∼32%) species shared a threonine in the same position. Analysis of seven C3 species with an antibody raised against a Thr78 phospho-peptide demonstrated that this position is phosphorylated in multiple genera. However, light-dependent dephosphorylation of Thr78 was observed only in Arabidopsis. Further, phosphorylation of Thr78 could not be detected in any of the four C4 grass species examined. The results suggest that despite conservation of Thr78 in Rubisco activase from a wide range of species, a regulatory role for phosphorylation at this site is more limited. This provides a case study for how variation in post-translational regulation can amplify functional divergence across the phylogeny of plants beyond what is explained by sequence variation in a metabolically important protein.
Data
Illinois Data Bank includes:
- Amino acid sequences
- Gene ID expression levels