Photoenzymatic Asymmetric Incorporation of Fluorinated Motifs into Olefins
Themes: Conversion
Keywords: Bioproducts, Catalysis
Citation
Li, M., Yuan, Y., Harrison, W., Zhang, Z., Zhao, H. June 11, 2024. “Asymmetric Photoenzymatic Incorporation of Fluorinated Motifs into Olefins.” Dryad. DOI: 10.5061/dryad.r2280gbm6.
Overview
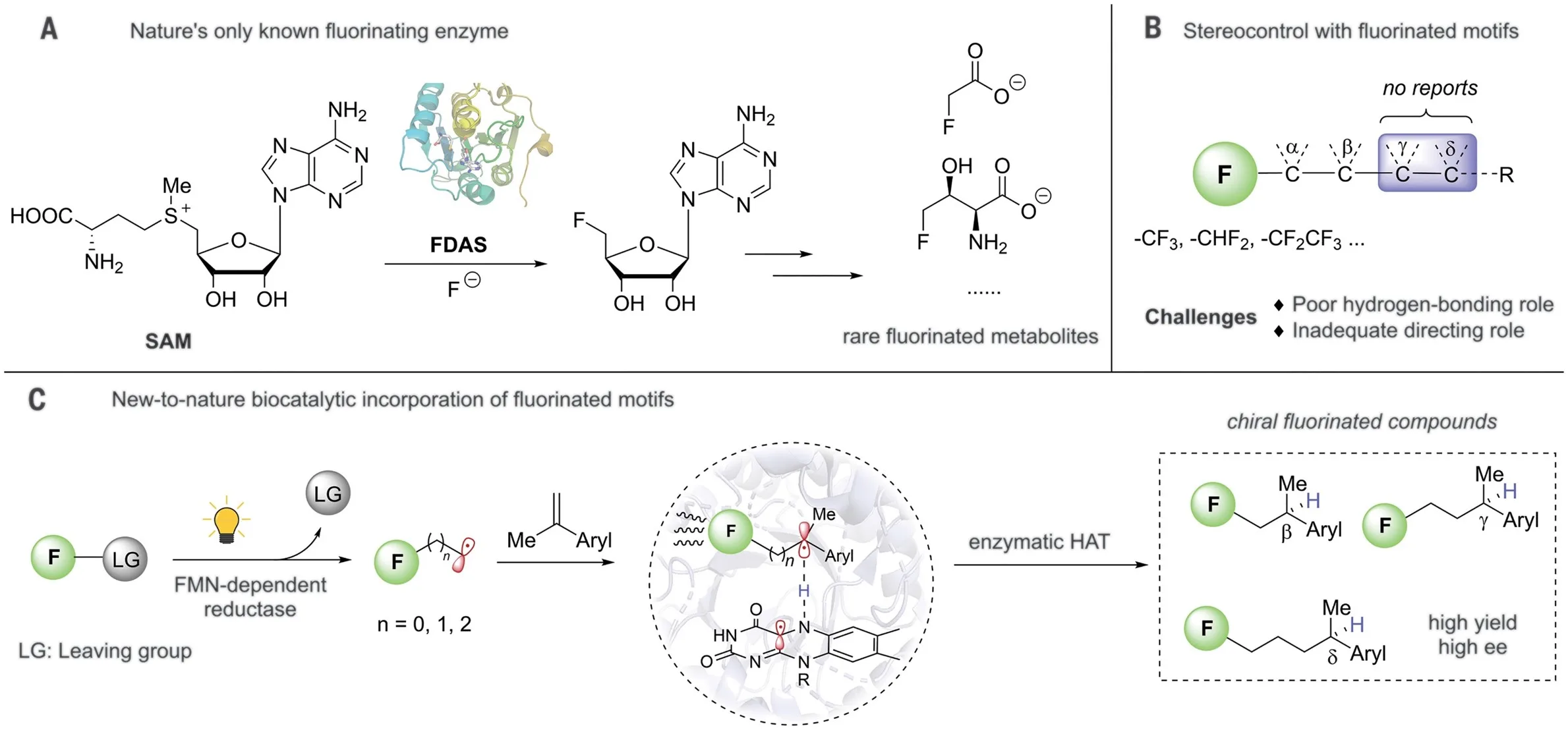
Enzymes capable of assimilating fluorinated feedstocks are scarce, which poses a challenge for the biosynthesis of fluorinated compounds used in pharmaceuticals, agrochemicals, and materials. We develop a photoenzymatic hydrofluoroalkylation that adeptly integrates fluorinated motifs into olefins. The photoinduced promiscuity of flavin-dependent ene-reductases enables generation of carbon-centered radicals from iodinated fluoroalkanes, which are directed by the photoenzyme to engage enantioselectively with olefins. This approach facilitates stereocontrol through interaction between a singular fluorinated unit and the enzyme, securing high enantioselectivity at β-, γ-, or δ-positions of fluorinated groups via enzymatic hydrogen atom transfer, a process notably challenging with conventional chemocatalysis. This work advances enzymatic strategies for integrating fluorinated chemical feedstocks and opens new avenues for asymmetric synthesis of fluorinated compounds.
This dataset supports the research published in “Photoenzymatic Asymmetric Incorporation of Fluorinated Motifs into Olefins” and contains all the necessary files for molecular docking and cluster modeling as described in the article and Supplementary Information. Molecular docking was completed using Autodock Vina, and cluster modeling was performed using Gaussian 16.
Data
Dryad – Includes pdbqt format of enzyme and ligand, docked result output file, molecular docking parameters configuration, cluster model coordinates