Multiallelic, Targeted Mutagenesis of Magnesium Chelatase with CRISPR-Cas9 Provides a Rapidly Scorable Phenotype in Highly Polyploid Sugarcane
Themes: Feedstock Production
Keywords: Genome Engineering, Genomics
Citation
Eid, A., Mohan, C., Sanchez, S., Wang, D., Altpeter, F. Feb. 23, 2021. “Multiallelic, targeted mutagenesis of magnesium chelatase with CRISPR-Cas9 provides a rapidly scorable phenotype in highly polyploid sugarcane.” NCBI.
Overview
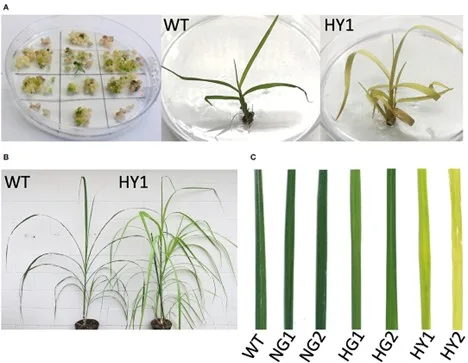
Genome editing with sequence-specific nucleases, such as clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9), is revolutionizing crop improvement. Developing efficient genome-editing protocols for highly polyploid crops, including sugarcane (x = 10-13), remains challenging due to the high level of genetic redundancy in these plants. Here, we report the efficient multiallelic editing of magnesium chelatase subunit I (MgCh) in sugarcane. Magnesium chelatase is a key enzyme for chlorophyll biosynthesis. CRISPR/Cas9-mediated targeted co-mutagenesis of 49 copies/alleles of magnesium chelatase was confirmed via Sanger sequencing of cloned PCR amplicons. This resulted in severely reduced chlorophyll contents, which was scorable at the time of plant regeneration in the tissue culture. Heat treatment following the delivery of genome editing reagents elevated the editing frequency two-fold and drastically promoted co-editing of multiple alleles, which proved necessary to create a phenotype that was visibly distinguishable from the wild type. Despite their yellow leaf color, the edited plants were established well in the soil and did not show noticeable growth retardation. This approach will facilitate the establishment of genome editing protocols for recalcitrant crops and support further optimization, including the evaluation of alternative RNA-guided nucleases to overcome the limitations of the protospacer adjacent motif (PAM) site or to develop novel delivery strategies for genome editing reagents.
Data
Nucleotide and SRA Experiments
Download (58 KB) includes:
- Primers
- SPAD leaf pigmentation data
- Phenotyping and genotyping summaries