Mitochondrial ATP Generation is More Proteome Efficient than Glycolysis
Themes: Conversion
Keywords: Metabolomics, Proteomics
Citation
Shen, Y., Dinh, H.V., Cruz, E.R., Chen, Z., Bartman, C.R., Xiao, T., Call, C.M., Ryseck, R.-P., Pratas, J., Weilandt, D., Baron, H., Subramanian, A., Fatma, Z., Wu, Z.-Y., Dwaraknath, S., Hendry, J.I., Tran, V.G., Yang, L., Yoshikuni, Y., Zhao, H., Maranas, C.D., Wuhr, M., Rabinowitz, J. April 25, 2024. Data from: “Mitochondrial ATP Generation is More Proteome Efficient than Glycolysis.” PRIDE.
Overview
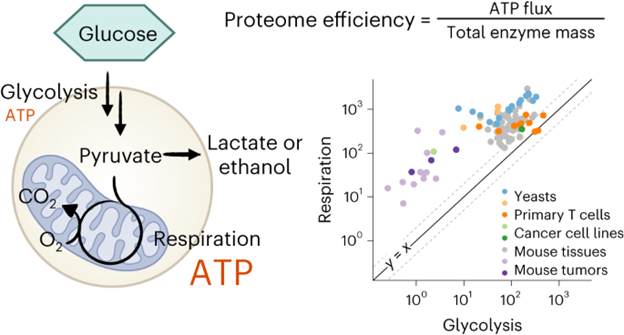
Metabolic efficiency profoundly influences organismal fitness. Heterotrophs, from yeast to mammals, derive usable energy primarily through glycolysis and respiration. While respiration is more energy-efficient, some cells favor glycolysis even when oxygen is available (aerobic glycolysis, Warburg effect). A leading explanation is that glycolysis is more efficient in terms of ATP production per unit mass of protein (i.e. faster). Through quantitative flux analysis and proteomics, we find however that mitochondrial respiration is actually more proteome-efficient than aerobic glycolysis. This is shown across yeasts, T cells, cancer cells, and tissues and tumors in vivo. Instead of aerobic glycolysis being valuable for fast ATP production, it correlates with high glycolytic protein expression, which is valuable for hypoxic growth. Aerobic glycolytic yeasts do not excel at aerobic growth, but outgrow respiratory cells in oxygen limitation. Thus, aerobic glycolysis emerges from cells maintaining a proteome conducive to both aerobic and hypoxic growth.
Data
PRIDE: Contains .raw and FTP files for S. cerevisiae
PRIDE: Contains .raw and FTP files for I. orientalis
GitHub: Supplementary files and R scripts regarding multi-omics data integration
GitHub: Supplementary files regarding 13C-metabolic flux analysis
Download (5.3 MB) includes:
- Metabolic fluxes of t-cell, cancer cell, mouse organ cell, and yeasts
- Proteome allocations and efficiencies
- Pyruvate ratios
- Glucose consumptions