Miscanthus sinensis Multi-Location Trial: Phenotypic Analysis, Genome-Wide Association, and Genomic Prediction
Themes: Feedstock Production
Keywords: Biomass Analytics, Genomics
Citation
Clark, L.V., Dwiyanti, M.S., Anzoua, K.G., Brummer, J.E., Ghimire, B.K., Głowacka, K., Hall, M., Heo, K., Jin, X., Lipka, A.E., Peng, J., Yamada, T., Yoo, J.H., Yu, C.Y., Zhao, H., Long, S.P., Sacks, E.J. March 25, 2019. Data from: “Genome-wide association and genomic prediction for biomass yield in a genetically diverse Miscanthus sinensis germplasm panel phenotyped at five locations in Asia and North America.” University of Illinois Urbana-Champaign. DOI: 10.13012/B2IDB-0790815_V3.
Overview
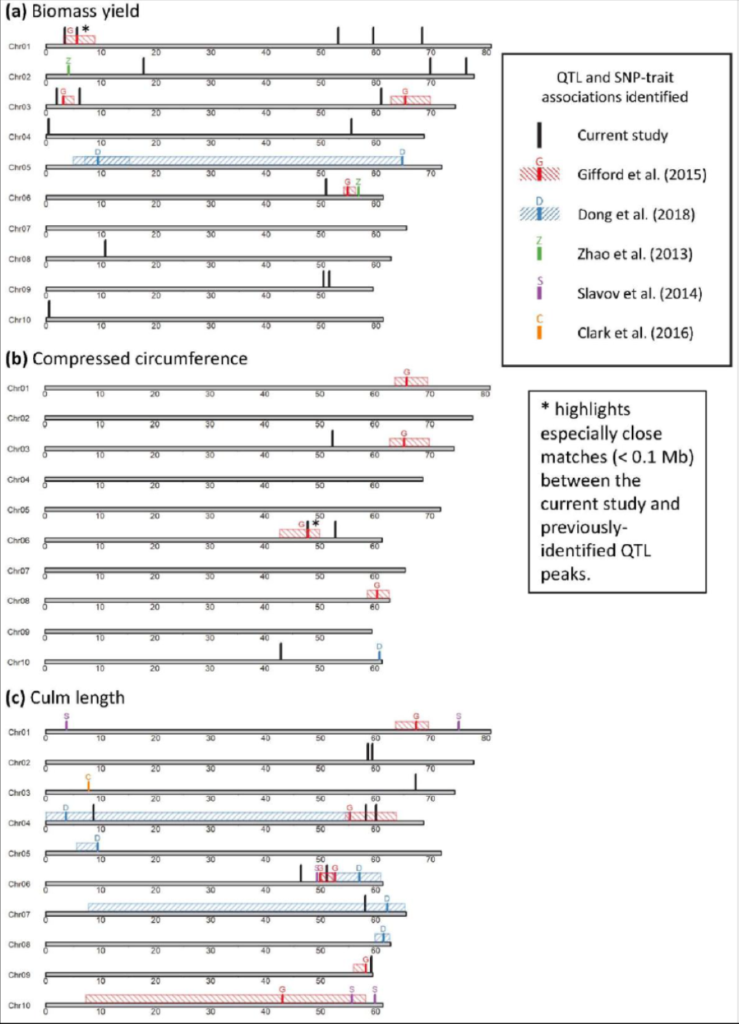
This dataset contains genotypic and phenotypic data, R scripts, and the results of analysis pertaining to a multi-location field trial of Miscanthus sinensis. Genome-wide association and genomic prediction were performed for biomass yield and 14 yield-component traits across six field trial locations in Asia and North America, using 46,177 single-nucleotide polymorphism (SNP) markers mined from restriction site-associated DNA sequencing (RAD-seq) and 568 M. sinensis accessions. Genomic regions and candidate genes were identified that can be used for breeding improved varieties of M. sinensis, which in turn will be used to generate new M. x giganteus clones for biomass.
Data
Illinois Data Bank – Genotypic and Phenotypic Datasets and Code
NCBI BioProject – Genetic Diversity Accession PRJNA207721
Download (1.4 MB) includes:
- Equations
- Residuals
- Metadata
- Sample info