Miscanthus sinensis Genome
Themes: Feedstock Production
Keywords: Genomics, RNA Sequencing, Transcriptomics
Citation
“Miscanthus sinensis v7.1,” Oct. 28, 2020. DOE-JGI, https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Msinensis_er
Overview
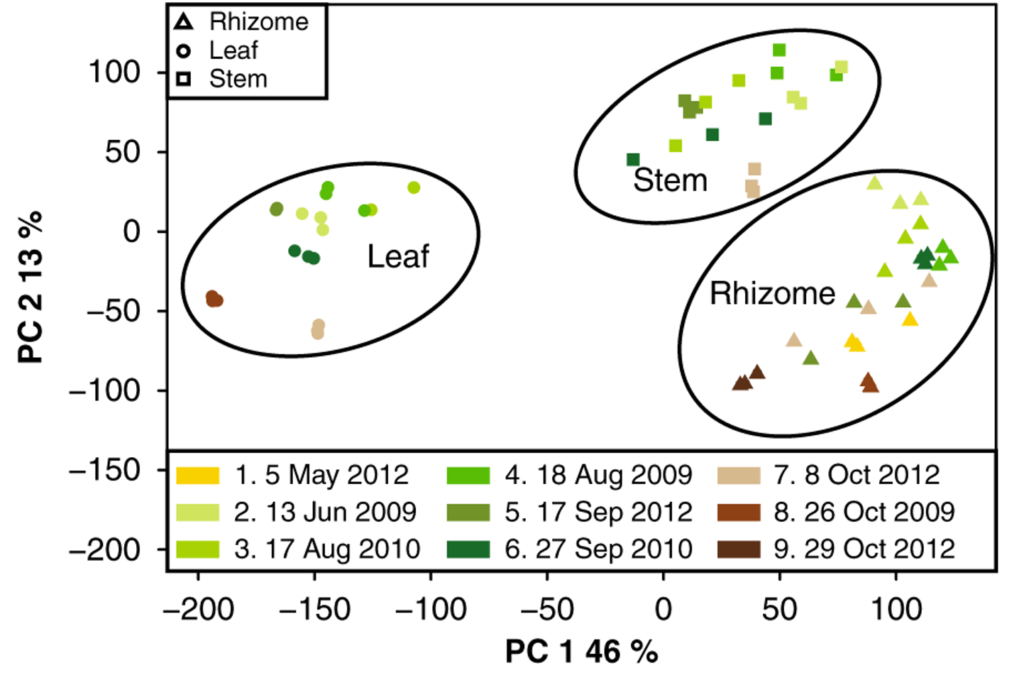
This is the first chromosome-scale assembly of Miscanthus sinensis doubled haploid DH1 (IGR-2011-001). The wild grass Miscanthus spp. is one of the world’s most widely adapted and productive plants. This chromosome-scale assembly will provide a foundational sequence resource for the Miscanthus genus, and an important link between the diploid Sorghum bicolor and the complex polyploid Saccharum species and provide a reference for the highly productive triploid M. x giganteus. Analysis of structural and regulatory changes among these genomes offer insights into the evolution of rhizome development, nutrient recycling, and self-incompatibility traits that contribute to highly efficient and sustainable biomass accumulation in a perennial temperate grass. Completion of the Miscanthus genome sequence represents a significant advance in both basic and applied plant genomics, creating a powerful system for comparative genomics among the Andropogoneae grasses that are of global importance as leading bioenergy feedstocks and food crops.
Data
Genomic reads for the M. sinensis DH1 genome assembly, PRJNA346689
RNASeq data in PRJNA575573 and SRP01f7791