Microbiome Differences in Sugarcane and Metabolically Engineered Oilcane Accessions and Their Implications for Bioenergy Production
Themes: Feedstock Production, Sustainability
Keywords: Biomass Analytics, Metabolomics
Citation
Yang, J., Sooksa-nguan, T., Kannan, B., Cano-Alfanar, S., Liu, H., Kent, A., Shanklin, J., Altpeter, F., Howe, A. March 30, 2023. “CABBI-oilcane 16S rRNA and ITS (greenhouse)” NIH – BioProject.
Overview
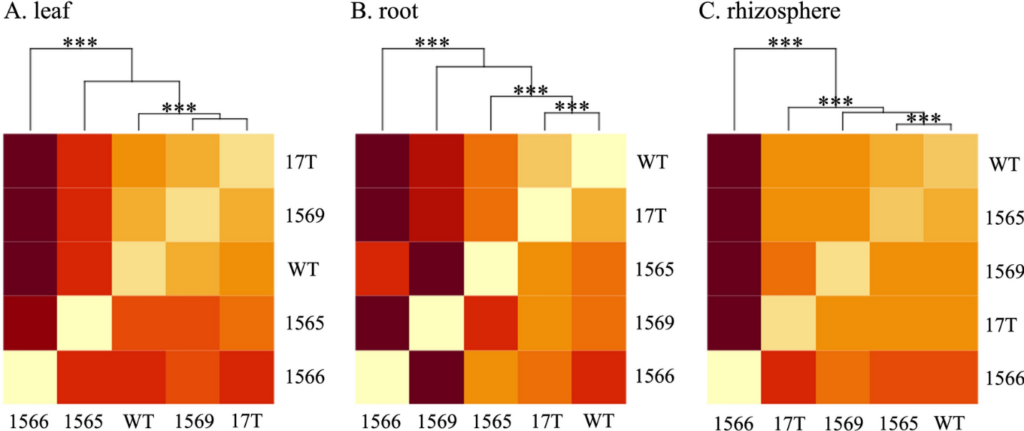
Oilcane is a metabolically engineered sugarcane (Saccharum spp. hybrid) that hyper-accumulates lipids in its vegetable biomass to provide an advanced feedstock for biodiesel production. The potential impact of hyper-accumulation of lipids in vegetable biomass on microbiomes and the consequences of altered microbiomes on plant growth and lipid accumulation have not been explored so far. Here, we explore differences in the microbiome structure of different oilcane accessions and non-modified sugarcane. 16S SSU rRNA and ITS rRNA amplicon sequencing were performed to compare the characteristics of the microbiome structure from different plant compartments (leaf, stem, root, rhizosphere, and bulk soil) of four greenhouse-grown oilcane accessions and non-modified sugarcane. Significant differences were only observed in the bacterial microbiomes. In leaf and stem microbiomes, more than 90% of the entire microbiome of non-modified sugarcane and oilcane was dominated by similar core taxa. Taxa associated with Proteobacteria led to differences in the non-modified sugarcane and oilcane microbiome structure. While differences were observed between multiple accessions, accession 1566 was notable in that it was consistently observed to differ in its microbial membership than other accessions and had the lowest abundance of taxa associated with plant-growth-promoting bacteria. Accession 1566 is also unique among oilcane accessions in that it has the highest constitutive expression of the WRI1 transgene. The WRI1 transcription factor is known to contribute to significant changes in the global gene expression profile, impacting plant fatty acid biosynthesis and photomorphogenesis. This study reveals for the first time that genetically modified oilcanes associate with distinct microbiomes. Our findings suggest potential relationships between core taxa, biomass yield, and TAG in oilcane accessions and support further research on the relationship between plant genotypes and their microbiomes.
Data
NIH – BioProject: Includes sequence data, BioSample
-
- Taxa Relative Abundance
- PERMANOVA
- Expression Analysis