Microbial Carbon Use Efficiency Predicted from Genome-Scale Metabolic Models
Themes: Sustainability
Keywords: Metabolomics, Modeling
Citation
Saifuddin, M., Bhatnagar, J.M., Segrè, D., Finzi, A.C. Aug. 8, 2019. Data from: “Microbial Carbon Use Efficiency Predicted from Genome-Scale Metabolic Models.” University of Illinois. 10.13012/B2IDB-6982180_V1.
Overview
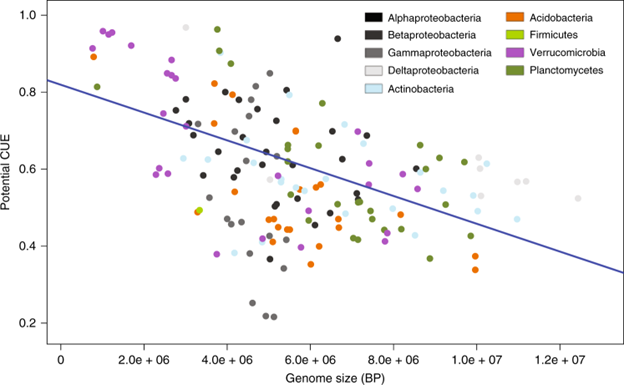
Respiration by soil bacteria and fungi is one of the largest fluxes of carbon (C) from the land surface. Although this flux is a direct product of microbial metabolism, controls over metabolism and their responses to global change are a major uncertainty in the global C cycle. Here, we explore an in silico approach to predict bacterial C-use efficiency (CUE) for over 200 species using genome-specific constraint-based metabolic modeling. We find that potential CUE averages 0.62 ± 0.17 with a range of 0.22 to 0.98 across taxa and phylogenetic structuring at the subphylum levels. Potential CUE is negatively correlated with genome size, while taxa with larger genomes are able to access a wider variety of C substrates. Incorporating the range of CUE values reported here into a next-generation model of soil biogeochemistry suggests that these differences in physiology across microbial taxa can feed back on soil-C cycling.
Data
Illinois Data Bank includes:
- CUE for BiGG Models
- CUE substrate limitation
- Contribution indices
- Regression summary