MALDI-MS Screening of Microbial Colonies With Isomer Resolution to Select Fatty Acid Desaturase Variants
Themes: Conversion
Keywords: Lipidomics, Mass Spectrometry
Citation
Choe, K., Jindra, M.A., Hubbard, S.C., Pfleger, B.F., Sweedler, J.V. May 3, 2023. “MALDI-MS Screening of Microbial Colonies with Isomer Resolution to Select Fatty Acid Desaturase Variants.” Analytical Biochemistry. DOI: 10.1016/j.ab.2023.115169.
Overview
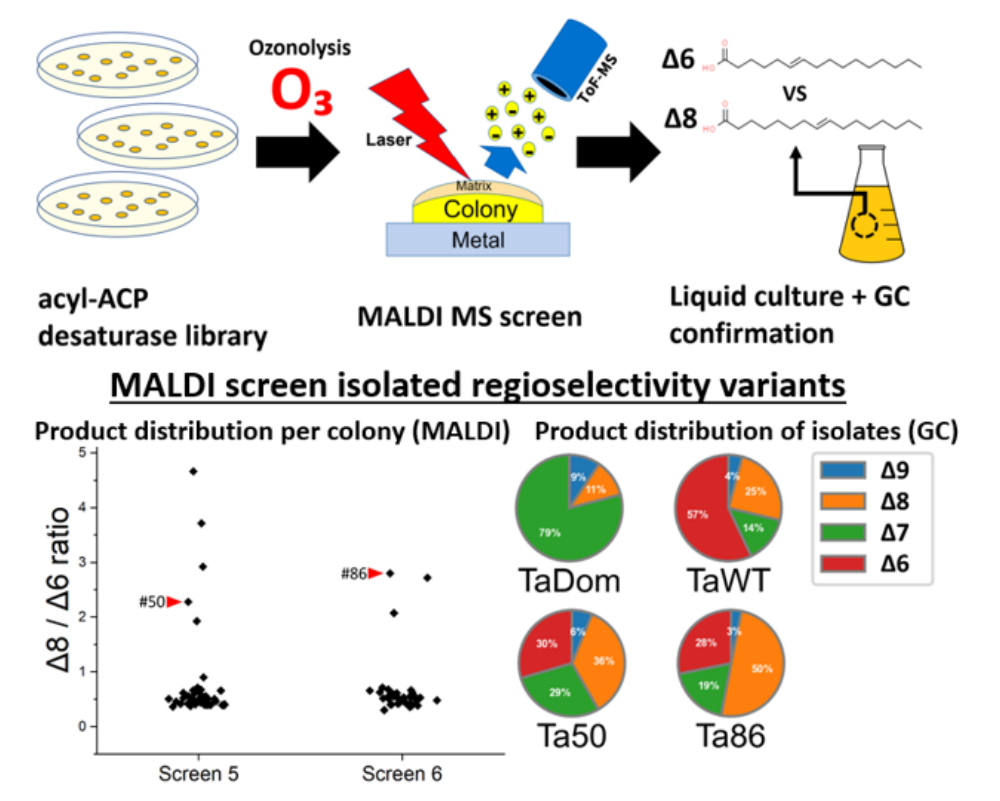
Creating controlled lipid unsaturation locations in oleochemicals can be a key to many bioengineered products. However, evaluating the effects of modifications to the acyl-ACP desaturase on lipid unsaturation is not currently amenable to high-throughput assays, limiting the scale of redesign efforts to <200 variants. Here, we report a rapid mass spectrometry (MS) assay for profiling the positions of double bonds on membrane lipids produced by Escherichia coli colonies after treatment with ozone gas. By MS measurement of the ozonolysis products of Δ6 and Δ8 isomers of membrane lipids from colonies expressing recombinant Thunbergia alata desaturase, we screened a randomly mutagenized library of the desaturase gene at 5 s per sample. Two variants with altered regiospecificity were isolated, indicated by an increase in 16:1 Δ8 proportion. We also demonstrated the ability of these desaturase variants to influence the membrane composition and fatty acid distribution of E. coli strains deficient in the native acyl-ACP desaturase gene, fabA. Finally, we used the fabA deficient chassis to concomitantly express a non-native acyl-ACP desaturase and a medium-chain thioesterase from Umbellularia californica, demonstrating production of only saturated free fatty acids.
Data
- MALDI data
- Desat dry LB plate data
- Membrane composition
- FFA production strains