macroMS
Themes: Conversion
Keywords: Mass Spectrometry, Software
Citation
Choe, K., Xue, P., Zhao, H., Sweedler, J.V. April 6, 2021. “macroMS: Image-Guided Analysis of Random Objects by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry.” Journal of the American Society of Mass Spectrometry. DOI: 10.1021/jasms.1c00013.
Overview
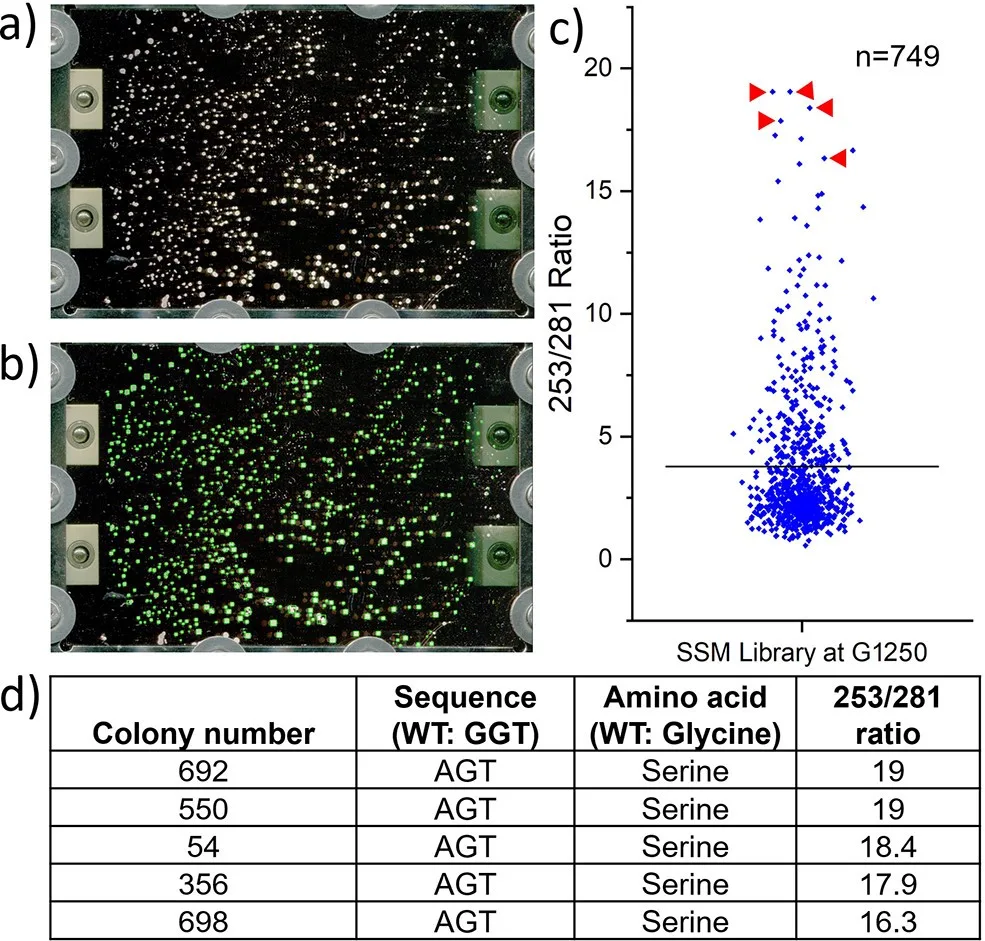
macroMS enables fast and simple workflow for image guided MALDI-ToF-MS analysis. Users upload images of ITO glasses or target plates holding samples, then macroMS finds target lists by image analysis. These targets are fed feed into MALDI-ToF-MS device for analysis. The macroMS includes limited data analysis for the generated data to aid finding analytes having compounds of interest. macroMS is freely available to academic and government laboratories for research purpose.
Materials requirement: 75 mm X 50 mm ITO glasses + Bruker TLC adapter (Bruker catalog #255595) or 100 mm X 75 mm X 1.1 mm ITO glasses + PAC II adapter (Bruker catalog # 221598). Alternative to the ITO glass, a 100 mm X 75 mm X 1.1 mm rectangular stainless plate with polished surface can be fabricated for the PAC II adapter. The system is built for and tested on ultrafleXtreme MALDI-TOF/TOF mass spectrometer.