Lost and Found: Rediscovering Microbiome-Associated Phenotypes that Reshape Agricultural Sustainability
Themes: Sustainability
Keywords: Agroecosystem, Metabolomics, Microbiome, RNA Sequencing
Citation
Favela, A., Kent, A.D., Sible, C.N., Flint-Garcia, S., Klimasmith, I.M., Mujjabi, C., Raglin, S.S., Below, F.E., Bohn, M.O. Jan. 1, 2026. Data from: “Lost and Found: Rediscovering Microbiome-Associated Phenotypes that Reshape Agricultural Sustainability.” Zenodo. DOI: 10.5281/zenodo.17656460.
Overview
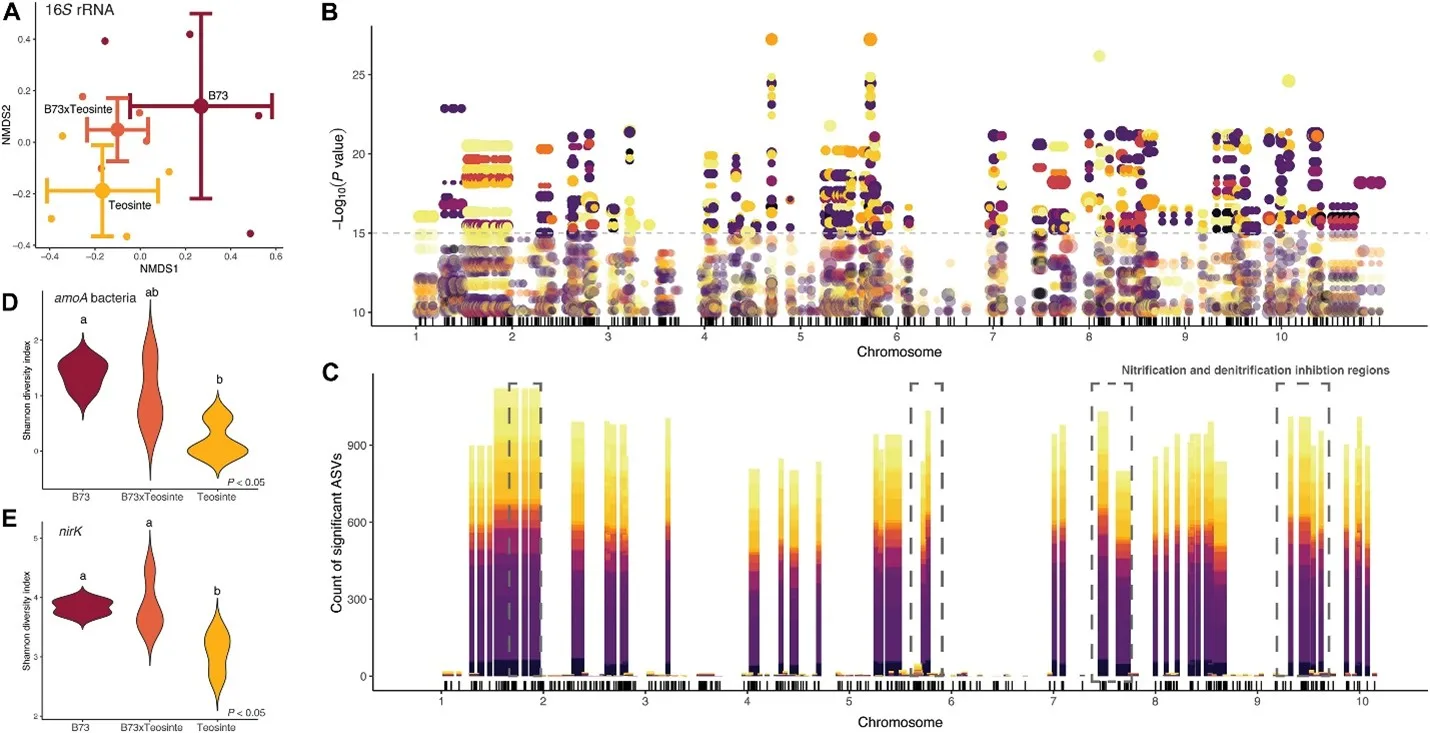
Code and data repository for NIL Manuscript. Documentation includes sequence processing examples and data analysis. Supplemental sequence processing and R statistical analysis for publication, which compares the microbiome of teosinte-B73 Near Isogenic Lines.
Sample Data
Amplicon sequence data for 16S rRNA genes, the fungal ITS2 region, and nitrogen-cycling functional genes are available through the NCBI Sequence Read Archive (SRA) under accession number PRJNA1042643 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1042643 ). Raw metabolomic data are available on Metabolomics Workbench, Project ID: PR002654. This study is available at the NIH Common Fund’s National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org where it has been assigned Study ID ST004211. The data can be accessed directly via its Project DOI: http://dx.doi.org/10.21228/M8KV8T.
Data
Zenodo: Code, functional data
NCBI – BioProject: Raw sample data
Metabolomics Workbench: Maize root exduate metabolomics data