Improving Precision and Accuracy of Genetic Mapping with Genotyping-by-Sequencing Data in Outcrossing Species
Themes: Feedstock Production
Keywords: Genetics, Genomics
Citation
LaBonte, N.R., Zerpa-Catanho, D.P., S, Liu, L. Xiao, Dong, H., L.V. Clark, E.J. Sacks. July 31, 2024. “Improving Precision and Accuracy of Genetic Mapping with Genotyping-by-Sequencing Data in Outcrossing Species.” University of Illinois Urbana-Champaign. DOI: 10.13012/B2IDB-1435220_V1.
Overview
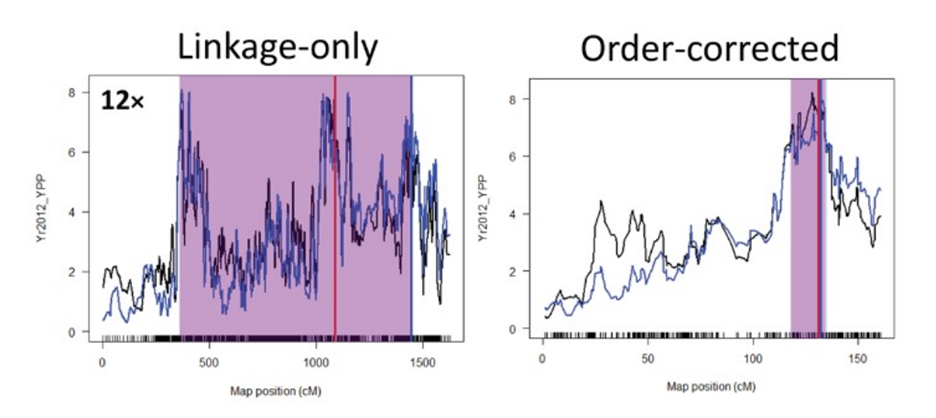
This dataset contains all data and supplementary materials from “Improving precision and accuracy of genetic mapping with genotyping-by-sequencing data in outcrossing species”. An Excel file a list of all QTLs and linkage group length (in cM) obtained with two different SNP-calling methods (Tassel-Uneak and Tassel-GBS), genetic map-construction method (linkage-only and reference order-corrected) and depth filters (12x, 20x, 30x and 40x) for genetic mapping of 18 biomass yield traits in a biparental Miscanthus sinensis population using RAD-Seq SNPs is provided as “Supplementary file 1”. A Perl script with the code for filtering VCF and HapMap-formatted data files is provided as “Supplementary file 2”. Phenotype data used for QTL mapping is provided as “Supplementary File 3”. A Perl script with the code for the simulation study is provided as “Supplementary file 4”.
Data
Illinois Data Bank – Includes QTLs, linkage group length, genetic map-construction method, depth filters, Perl scripts, phenotype data
NIH NCBI BioProject – Includes Miscanthus sinensis sample data