GNPS – Decompartmentalization of the Yeast Mitochondrial Metabolism to Improve Chemical Production in Issatchenkia orientalis
Themes: Conversion, Sustainability
Keywords: Bioproducts, Metabolic Engineering
Citation
Tran, V.G., Tan, S.-I., Xu, H., Weilandt, D.R., Li, X., Bhagwat, S.S., Zhu, Z., Guest, J.S., Rabinowitz, J.D., Zhao, H. Aug. 2, 2025. ““GNPS – Decompartmentalization of the yeast mitochondrial metabolism to improve chemical production in Issatchenkia orientalis.” CCMS – MassIVE. DOI: 10.25345/C5TX35K3J.
Overview
This dataset contains the raw LC-MS data of metabolites extracted from I. orientalis strains. [doi:10.25345/C5TX35K3J] [dataset license: CC0 1.0 Universal (CC0 1.0)]
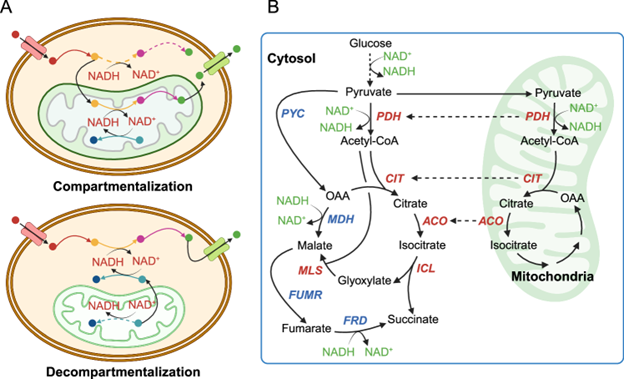
Microbial production of chemicals may suffer from inadequate cofactor provision, a challenge further exacerbated in yeasts due to compartmentalized cofactor metabolism. Here, we perform cofactor engineering through the decompartmentalization of mitochondrial metabolism to improve succinic acid (SA) production in Issatchenkia orientalis. We localize the reducing equivalents of mitochondrial NADH to the cytosol through cytosolic expression of its pyruvate dehydrogenase (PDH) complex and couple a reductive tricarboxylic acid pathway with a glyoxylate shunt, partially bypassing an NADH-dependent malate dehydrogenase to conserve NADH. Cytosolic SA production reaches a titer of 104 g/L and a yield of 0.85 g/g glucose, surpassing the yield of 0.66 g/g glucose constrained by cytosolic NADH availability. Additionally, expressing cytosolic PDH, we expand our I. orientalis platform to enhance acetyl-CoA-derived citramalic acid and triacetic acid lactone production by 1.22- and 4.35-fold, respectively. Our work establishes I. orientalis as a versatile platform to produce markedly reduced and acetyl-CoA-derived chemicals.
Data
CCMS – MassIVE: includes raw LC-MS data of metabolites extracted from I. orientalis strains
Illinois Data Bank: includes supplemental information, fermentation conditions and profiles, titers, strains, primers, plasmids, and reactions.