Enhancing Lipid Production in Plant Cells through Automated High-Throughput Genome Engineering and Phenotyping
Themes: Conversion, Feedstock Production
Keywords: AI/ML, Genome Engineering, Metabolic Engineering, Phenotyping
Citation
Dong, J., Crossbow, S.W., Lane, S.T., Castro, D.C., Blanford, J., Zhou, S., Park, K., Burgess, S., Root, M., Cahoon, E., Shanklin, J., Sweedler, J.V., Zhao, H., Hudson, M.E. Feb. 3, 2025. Data from: “Enhancing Lipid Production in Plant Cells through Automated High-Throughput Genome Editing and Phenotyping.” University of Illinois. DOI: 10.13012/B2IDB-5553768_V1.
Overview
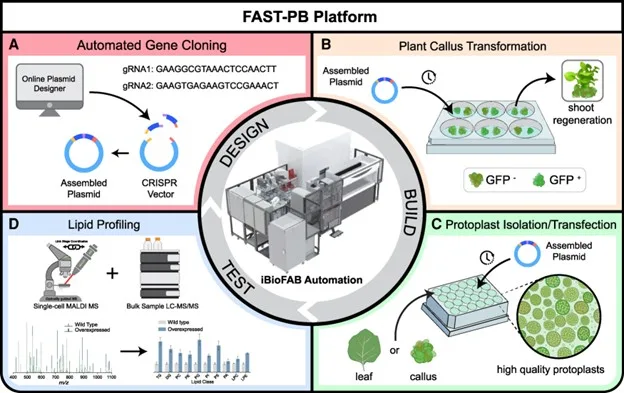
Plant bioengineering is a time-consuming and labor-intensive process with no guarantee of achieving desired traits. Here, we present a fast, automated, scalable, high-throughput pipeline for plant bioengineering (FAST-PB) in maize (Zea mays) and Nicotiana benthamiana. FAST-PB enables genome editing and product characterization by integrating automated biofoundry engineering of callus and protoplast cells with single-cell matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS). We first demonstrated that FAST-PB could streamline Golden Gate cloning, with the capacity to construct 96 vectors in parallel. Using FAST-PB in protoplasts, we found that PEG2050 increased transfection efficiency by over 45%. For proof-of-concept, we established a reporter-gene-free method for CRISPR editing and phenotyping via mutation of high chlorophyll fluorescence 136. We show that diverse lipids were enhanced up to 6-fold using CRISPR activation of lipid controlling genes. In callus cells, an automated transformation platform was employed to regenerate plants with enhanced lipid traits through introducing multigene cassettes. Lastly, FAST-PB enabled high-throughput single-cell lipid profiling by integrating MALDI-MS with the biofoundry, protoplast, and callus cells, differentiating engineered and unengineered cells using single-cell lipidomics. These innovations massively increase the throughput of synthetic biology, genome editing, and metabolic engineering and change what is possible using single-cell metabolomics in plants.
Data
Benchling: pMDC43
Benchling: pMDC43-OWD
Benchling: pPTN1586 – pGBSbM14n-2
Benchling: pZP212
- Plasmid CRISPR knockout/activation
- MALDI-MS mass match assignments
- Time/cost savings
- gRNAs/primers