Discovery, Characterization, and Application of Chromosomal Integration Sites for Stable Heterologous Gene Expression in Rhodotorula toruloides
Themes: Conversion
Keywords: Metabolic Engineering, Software, Transcriptomics
Citation
Xu, H., Shi, L., Boob, A.G., Park, W., Tan, S.I., Tran, V.G., Schultz, J.C., Zhao, H. Feb. 14, 2025. Data from: “Discovery, Characterization, and Application of Chromosomal Integration Sites for Stable Heterologous Gene Expression in Rhodotorula toruloides.” GitHub.
Overview
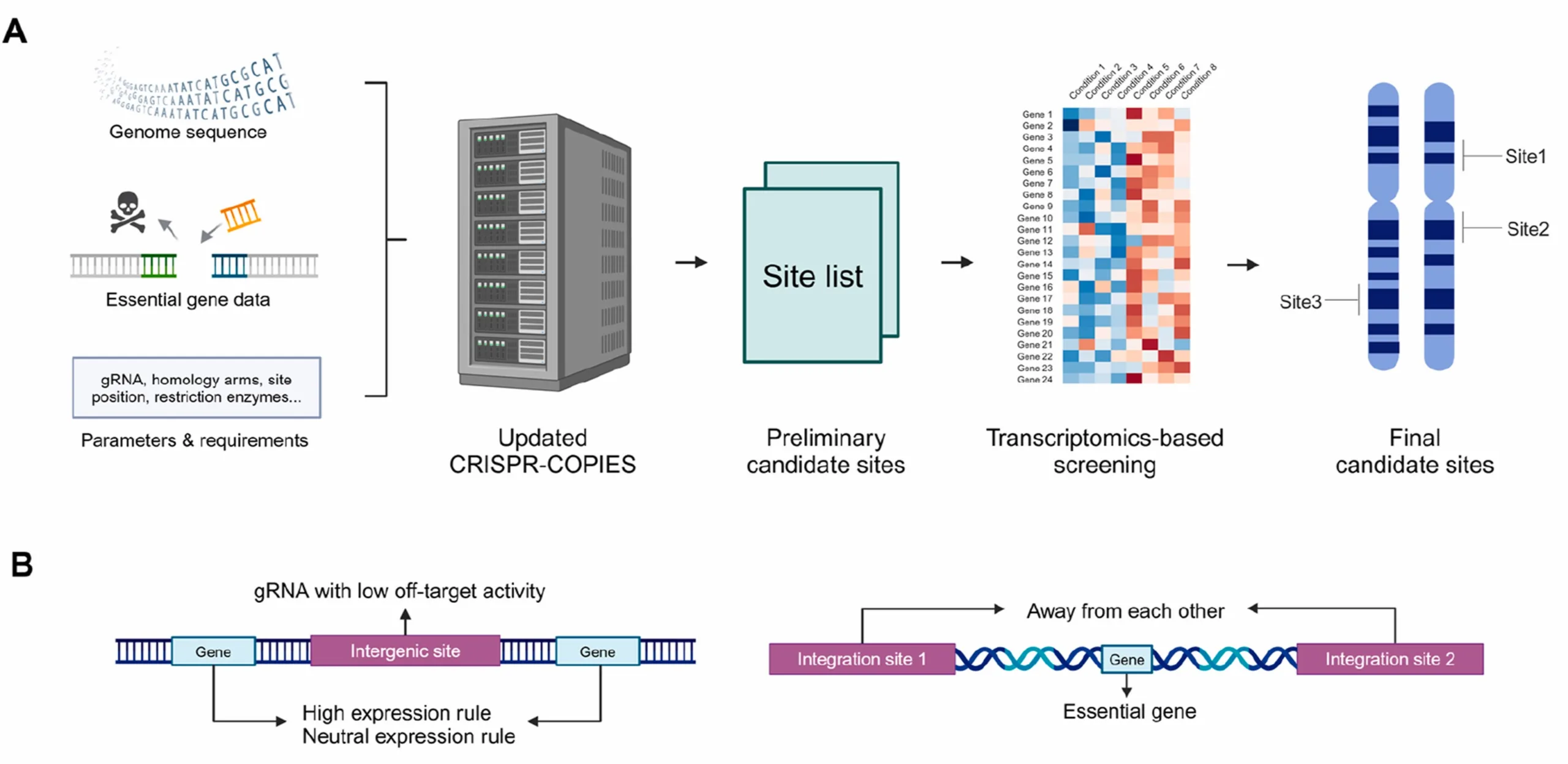
Rhodotorula toruloides is a non-model, oleaginous yeast uniquely suited to produce acetyl-CoA-derived chemicals. However, the lack of well-characterized genomic integration sites has impeded the metabolic engineering of this organism. Here we report a set of computationally predicted and experimentally validated chromosomal integration sites in R. toruloides. We first implemented an in silico platform by integrating essential gene information and transcriptomic data to identify candidate sites that meet stringent criteria. We then conducted a full experimental characterization of these sites, assessing integration efficiency, gene expression levels, impact on cell growth, and long-term expression stability. Among the identified sites, 12 exhibited integration efficiencies of 50% or higher, making them sufficient for most metabolic engineering applications. Using selected high-efficiency sites, we achieved simultaneous double and triple integrations and efficiently integrated long functional pathways (up to 14.7 kb). Additionally, we developed a new inducible marker recycling system that allows multiple rounds of integration at our characterized sites. We validated this system by performing five sequential rounds of GFP integration and three sequential rounds of MaFAR integration for fatty alcohol production, demonstrating, for the first time, precise gene copy number tuning in R. toruloides. These characterized integration sites should significantly advance metabolic engineering efforts and future genetic tool development in R. toruloides.
Data
GitHub: Python scripts, genome, feature table, protein sequences
Illinois Data Bank includes:
- CRISPR-COPIES parameters
- Integration sites
- Strains/plasmids/primers