Development of a CRISPR/Cas9-Based Tool for Gene Deletion in Issatchenkia orientalis
Themes: Conversion
Keywords: Genome Engineering, Genomics, Transcriptomics
Citation
Tran, V.G., Cao, M., Fatma, Z., Song, X., Zhao, H. June 26, 2019. Data from: “Development of a CRISPR/Cas9-Based Tool for Gene Deletion in Issatchenkia orientalis.” University of Illinois Urbana-Champaign. DOI:10.13012/B2IDB-4661029_V1.
Overview
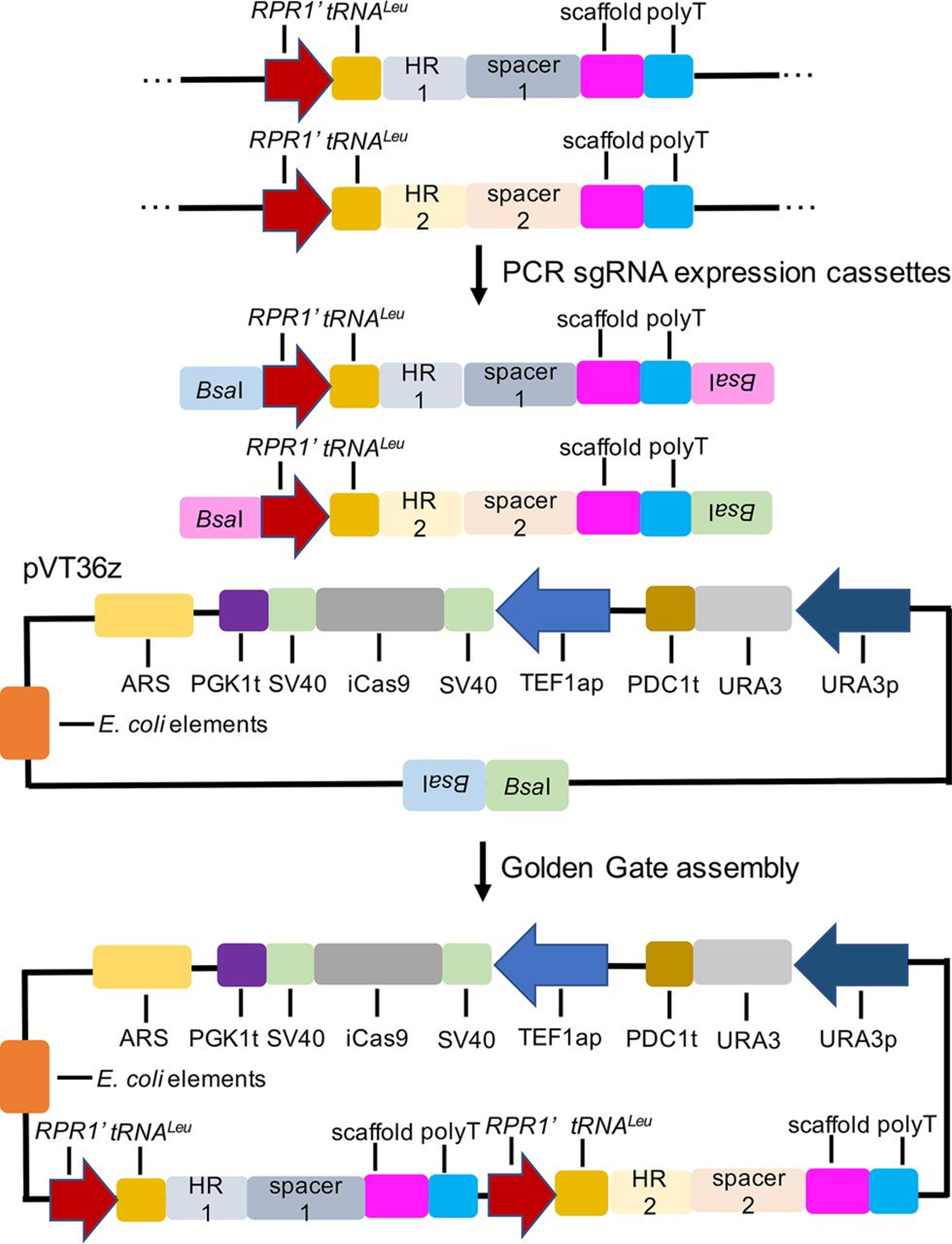
The nonconventional yeast Issatchenkia orientalis has emerged as a potential platform microorganism for production of organic acids due to its ability to grow robustly under highly acidic conditions. However, lack of efficient genetic tools remains a major bottleneck in metabolic engineering of this organism. Here we report that the autonomously replicating sequence (ARS) from Saccharomyces cerevisiae (ScARS) was functional for plasmid replication in I. orientalis, and the resulting episomal plasmid enabled efficient genome editing by the CRISPR/Cas9 system. The optimized CRISPR/Cas9-based system employed a fusion RPR1′-tRNA promoter for single guide RNA (sgRNA) expression and could attain greater than 97% gene disruption efficiency for various gene targets. Additionally, we demonstrated multiplexed gene deletion with disruption efficiencies of 90% and 47% for double gene and triple gene knockouts, respectively. This genome editing tool can be used for rapid strain development and metabolic engineering of this organism for production of biofuels and chemicals.
Data
Illinois Data Bank includes:
- Strains and plasmids
- Disruption efficiencies
- Primers, gBlock, and RNA sequences