
Plasmids have extensive use in basic and applied biology. These small, circular DNA molecules are used by scientists to introduce new genes into a target organism. Well known for their applications in the production of therapeutic proteins like insulin, plasmids are broadly used in the large-scale production of many bioproducts.
However, designing and constructing plasmids remains one of the most time-consuming and labor-intensive steps in biology research.
To address this, Behnam Enghiad, Pu Xue, and other University of Illinois Urbana-Champaign researchers at the Center for Advanced Bioenergy and Bioproducts Innovation (CABBI) have developed a versatile and automated platform for plasmid design and construction called PlasmidMaker. Their work was recently published in Nature Communications.
Creating a plasmid starts with design. To aid in this design process, PlasmidMaker has a user-friendly web interface with which researchers can intuitively visualize and assemble the perfect plasmid for their needs.
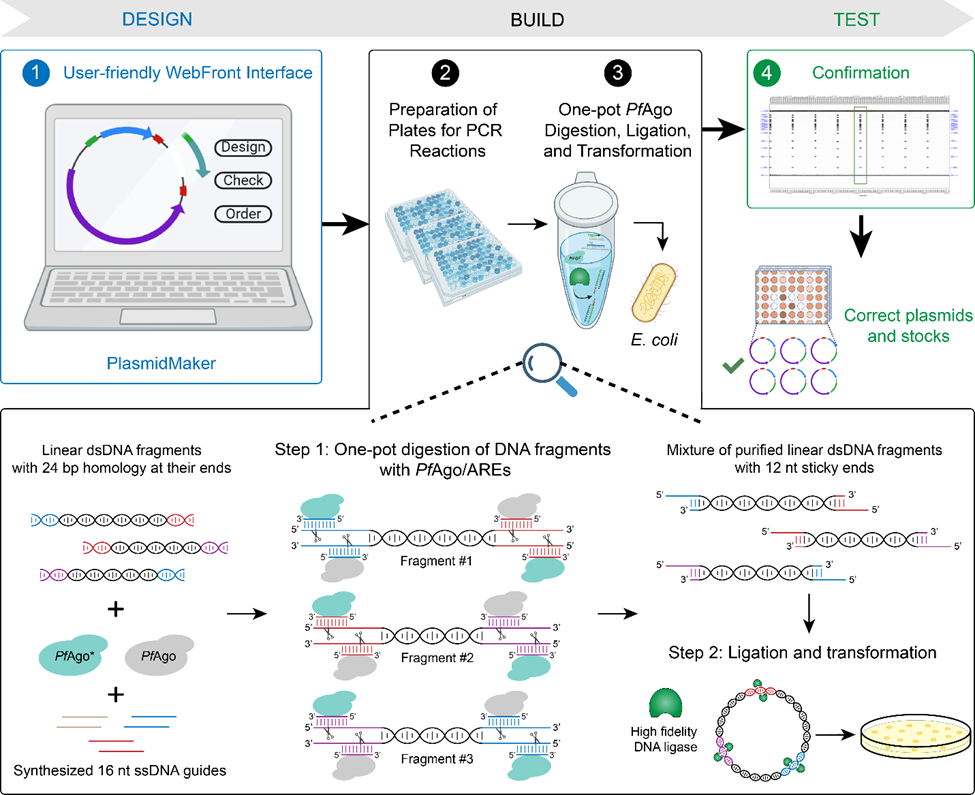
Once the plasmid has been designed, it is submitted to the PlasmidMaker team, and an order for the plasmid is placed at the Illinois Biological Foundry for Advanced Biomanufacturing (iBioFAB), where the plasmid will be built. iBioFAB, located at the Carl R. Woese Institute for Genomic Biology (IGB) on the U of I campus, is a fully integrated computational and physical infrastructure that supports rapid fabrication, quality control, and analysis of genetic constructs. It features a central robotic arm that transfers labware between instruments that perform distinct operations like pipetting, incubation, or thermocycling.
The plasmid build process is automated: samples are prepared through polymerase chain reaction (PCR) and purification, the DNA sequence is assembled and transformed, and the plasmids are confirmed and frozen, all with little human involvement.
In addition to the automation and precision afforded by iBioFAB, the PlasmidMaker platform also pioneers a new highly flexible method for assembling multiple DNA fragments into a plasmid using Pyrococcus furiosus Argonaute (PfAgo)-based artificial restriction enzymes (AREs).
Restriction enzymes have long been used in plasmid construction, as they can cleave DNA molecules at specific sequences of bases, called recognition sequences. However, these recognition sequences are usually short, making them hard to work with. A short sequence is likely to occur multiple times in a DNA molecule, in which case the restriction enzyme would make too many cuts.
“In previous DNA assembly methods, it would often be hard to find the right restriction enzymes that can cut the plasmid and replace the DNA fragments,” said Huimin Zhao, co-author and the Steven L. Miller Chair of Chemical and Biomolecular Engineering (ChBE) at Illinois. “The PfAgo-based AREs offer greater flexibility and precision, as they can be programmed to seek out longer recognition sequences at virtually any site.”
With all the improvements it brings to the table, the team members at CABBI, one of four U.S. Department of Energy-funded Bioenergy Research Centers across the United States, hope that PlasmidMaker will accelerate the development of synthetic biology for biotechnological applications.
“This tool will be available to CABBI researchers, and we want to eventually make it available to all researchers at the other three Bioenergy Research Centers,” Zhao said. “If things go well, we hope to make it available to all researchers everywhere.”
The manuscript’s other co-authors are Nilmani Singh, CABBI Automation Engineer; Aashutosh Girish Boob and Chengyou Shi, CABBI graduate students in ChBE; Vassily Andrew Petrov, CABBI Software Engineer; Roy Liu, CABBI undergraduate student in Computer Engineering; Siddhartha Suryanarayana Peri, CABBI undergraduate student in ChBE; Stephan Thomas Lane, CABBI iBioFAB Manager; and Emily Danielle Gaither, former CABBI iBioFAB Technician.
Read the full journal article in Nature Communications >>>
— Article by CABBI Communications Specialist April Wendling