rtRBA: R. toruloides Resource Balance Analysis Model
Themes: Conversion
Keywords: Gene Expression, Metabolomics, Modeling, Proteomics
Citation
Mooney, E.J., Suthers, P.F., Schroeder, W.L., Dinh, H.V., Li, X., Shen, Y., Xiao, T., Call, C.M., Baron, H., Subramanian, A.M., Weilandt, D.R., Keber, F.C., Wühr, M., Rabinowitz, J.D., Maranas, C.D. August 12, 2025. “rtRBA: R. toruloides Resource Balance Analysis Model.” GitHub.
Overview
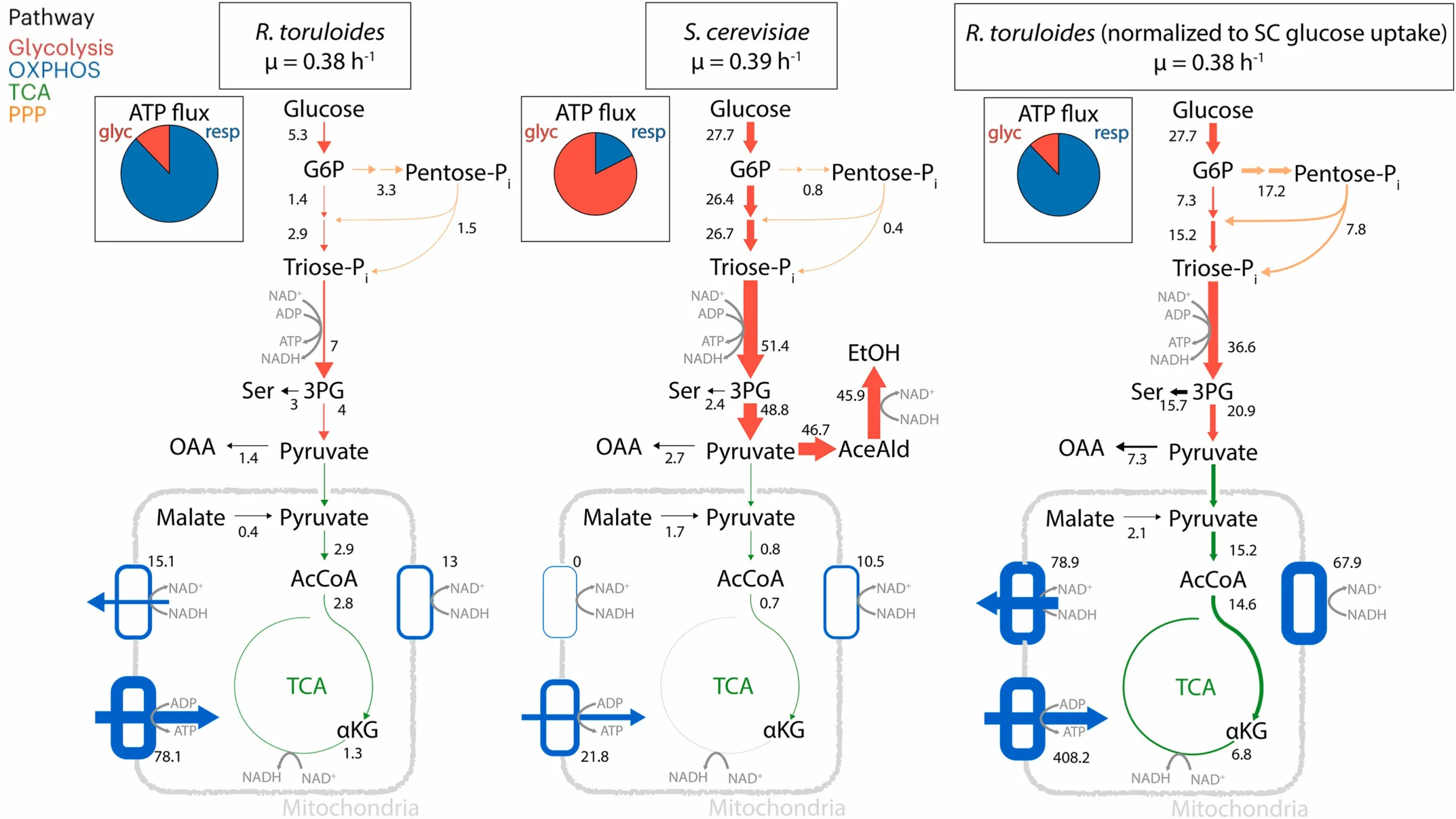
This repository provides resources and model files for the genome-scale resource balance analysis (RBA) model rtRBA. Eric J. Mooney and Costas D. Maranas
Additional info is present in the publication for this model.
Program and packages requirements for the provided software implementation: Python 3 (plus packages: cobra, pandas, numpy, matplotlib, jupyter, scikit-learn, cplex (compiled from installed CPLEX linear programming solver), and all automatically-installed associated dependencies to those already listed) and GAMS with (“built-in”) Soplex solver. (Tested on Python 3.6 and GAMS 39.1.0. Python package installation could be done through “pip” and could take up to 1 hour) gsm_custom_functions.py contains many useful functions, and may be easier to use if you move it into your $PYTHONPATH directory.”
Data
GitHub: Python scripts, model resources