Efficient Mutagenesis and Genotyping of Maize Inbreds Using Biolistics, Multiplex CRISPR/Cas9 Editing, and Indel-Selective PCR
Themes: Feedstock Production
Keywords: Gene Editing, Genome Engineering, Genotyping
Citation
Rai, M.N., Rhodes, B.H., Jinga, S., Kanchupati, P., Ross, E.H., Carlson, S.R., Moose, S.P. March 28, 2025. “Genome seq of edited H99 callus in maize.” NIH – NCBI.
Overview
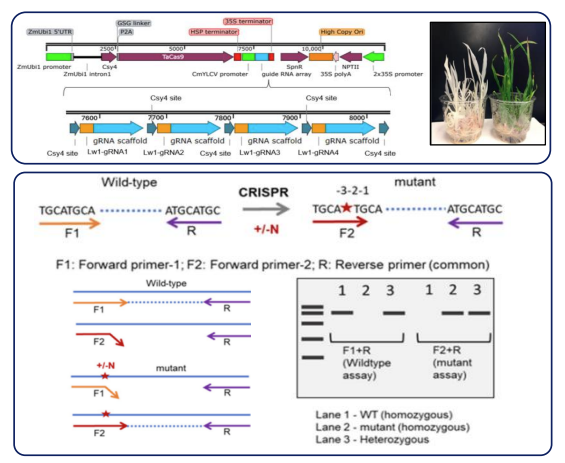
To identify and quantify any off-target mutations created during the NRT1.1 CRISPR/Cas9 experiment, genomic DNA was extracted from NRT1.1-E1 callus tissue for deep sequencing. Genomic library preparation was conducted using the Nextera DNA Flex Library Prep Kit (Illumina Cat #: 20018704) according to the manufacturers protocol and sequencing was performed on an Illumina NovaSeq6000.
Data
NIH – NCBI: SRX26347042: Genome sequence of edited H99 callus in maize
Illinois Data Bank includes:
- Primers
- Transformation efficiency
- CRISPR-Cas9 editing outcomes
- Alignment results